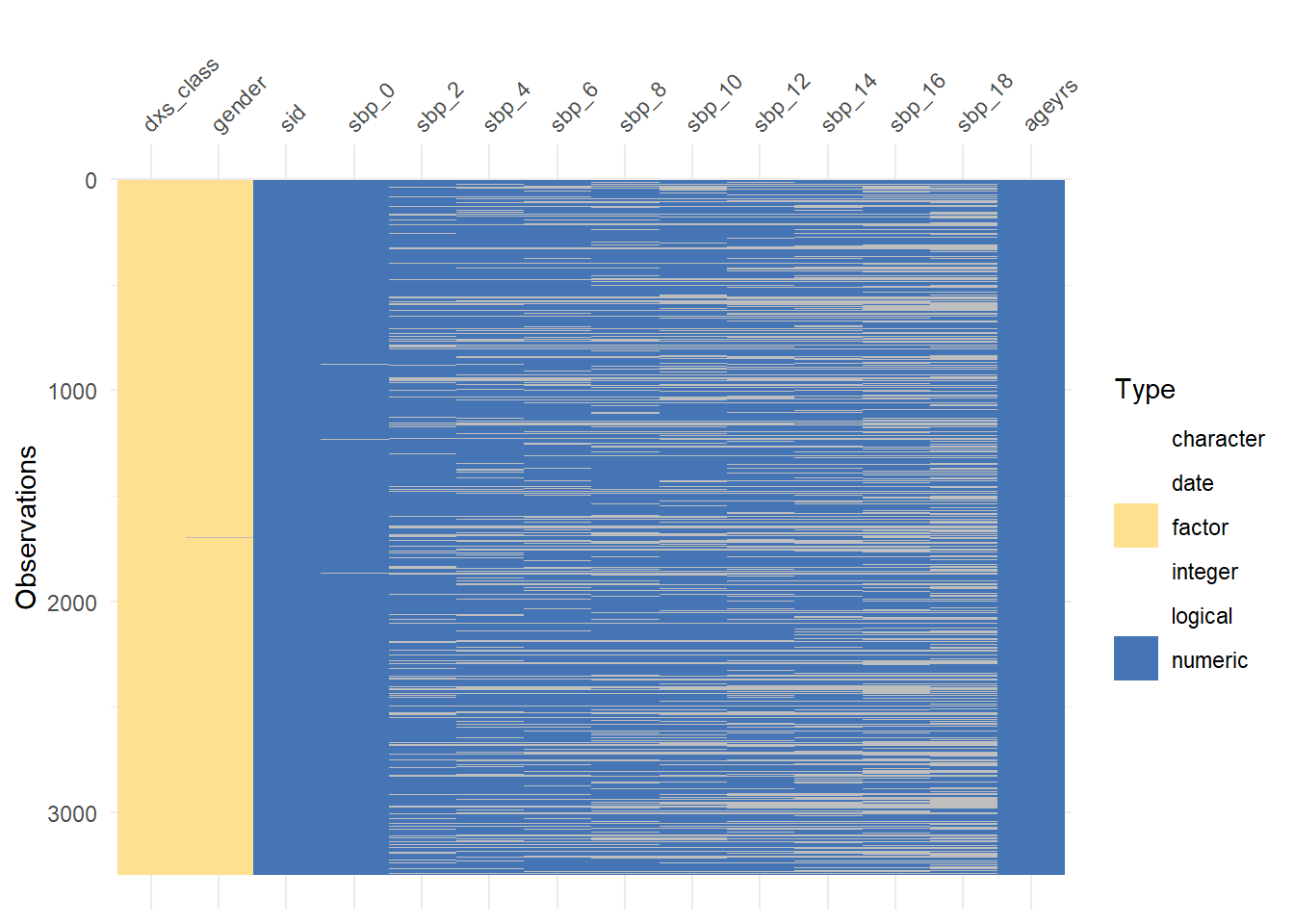
Rows: 3,296
Columns: 14
$ sid <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
$ dxs_class <fct> HPT, DM+HPT, HPT, HPT, HPT, DM+HPT, DM+HPT, DM, HPT, HPT, HP…
$ sbp_0 <dbl> 139, 155, 109, 130, 124, 140, 137, 160, 153, 135, 112, 137, …
$ sbp_2 <dbl> 124, NA, 123, NA, 120, 114, 135, 130, 218, 130, 118, 150, 14…
$ sbp_4 <dbl> 130, NA, 109, NA, 146, 163, 132, NA, NA, 118, 112, 130, 138,…
$ sbp_6 <dbl> 130, NA, 126, NA, 144, 117, 147, NA, NA, 150, 141, 112, 120,…
$ sbp_8 <dbl> 104, NA, 108, NA, 157, 124, 130, NA, NA, NA, 120, 129, 148, …
$ sbp_10 <dbl> 129, NA, 115, NA, 123, 121, NA, NA, 218, 127, NA, 148, 143, …
$ sbp_12 <dbl> 80, NA, 115, NA, 120, 128, 124, NA, NA, NA, NA, 142, NA, 116…
$ sbp_14 <dbl> 129, NA, 122, NA, 131, 119, 142, NA, NA, 123, NA, 131, NA, 1…
$ sbp_16 <dbl> 126, NA, 131, NA, 120, 100, 144, NA, NA, 149, 96, 119, NA, 2…
$ sbp_18 <dbl> 135, NA, 102, NA, 123, 127, 128, NA, NA, 132, 119, 119, 167,…
$ ageyrs <dbl> 75, 60, 62, 70, 72, 56, 51, 73, 61, 59, 75, 53, 43, 64, 70, …
$ gender <fct> Male, Male, Male, Male, Male, Male, Male, Male, Female, Male…