Code
df <-
data.frame(
dose = c("D0.5", "D1", "D2"),
len = c(4.2, 10, 29.5))
df dose len
1 D0.5 4.2
2 D1 10.0
3 D2 29.5To start, we create fictitious data
df <-
data.frame(
dose = c("D0.5", "D1", "D2"),
len = c(4.2, 10, 29.5))
df dose len
1 D0.5 4.2
2 D1 10.0
3 D2 29.5Next, we make the basic Barplot
p <-
df %>%
ggplot(aes(x = dose, y = len))
p +
geom_bar(stat = "identity")+
labs(
x = "Dose (mg)",
y = 'Tooth Length (mm)',
title = "Distribution of Tooth Lenghts for the variuos dosee")+
theme_classic()+
coord_cartesian(expand = FALSE)
Next we flip the barplot horizontal, change the size of the bar width and change the theme
p +
geom_bar(
stat = "identity",
width = 0.8,
color = "blue",
fill = "grey90") +
coord_flip() +
theme_bw()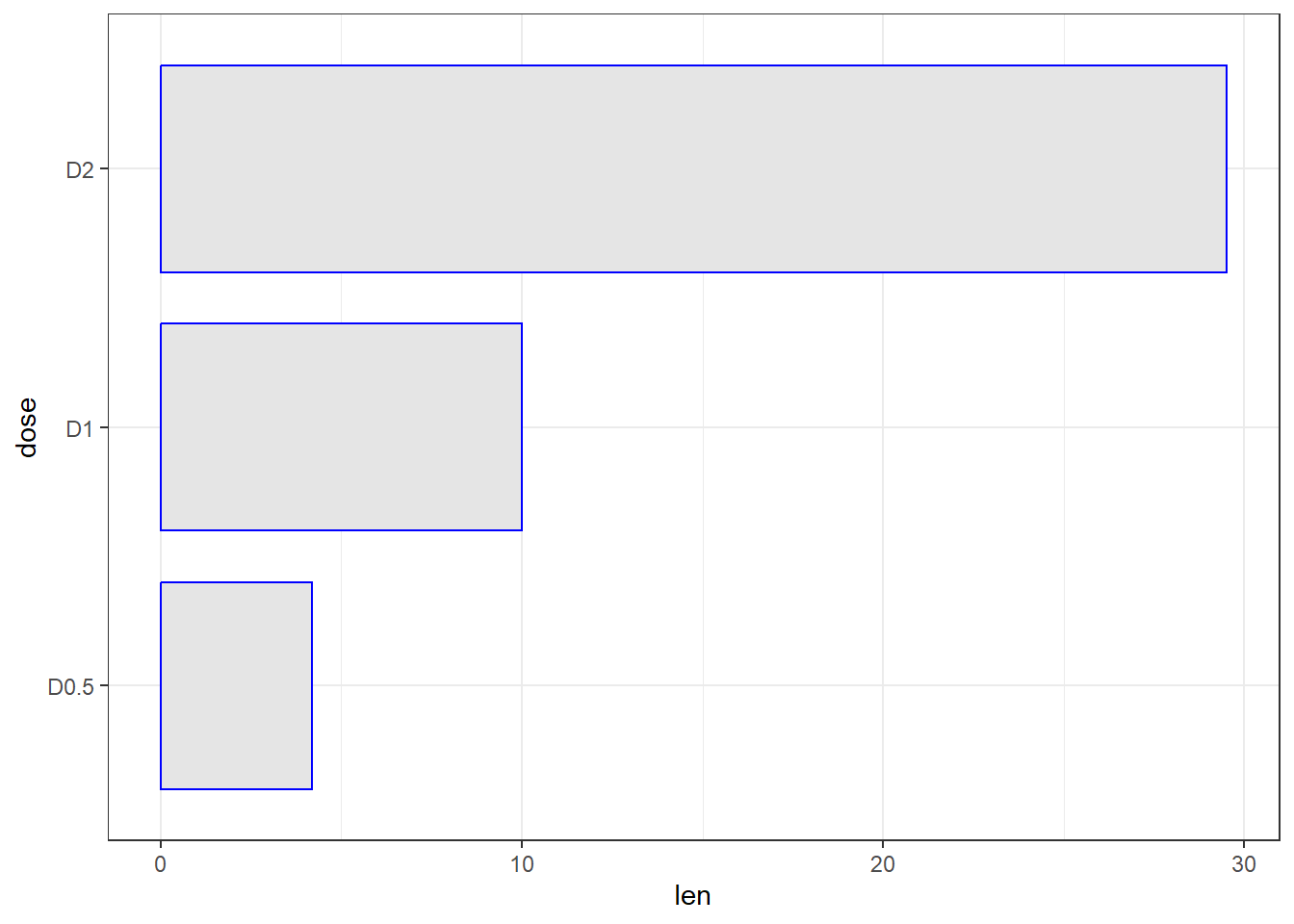
Next we limit the observations to just two
p +
geom_bar(
stat = "identity",
width = 0.8,
color = "black",
fill = "steelblue") +
scale_x_discrete(limits=c("D0.5", "D2"))Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_bar()`).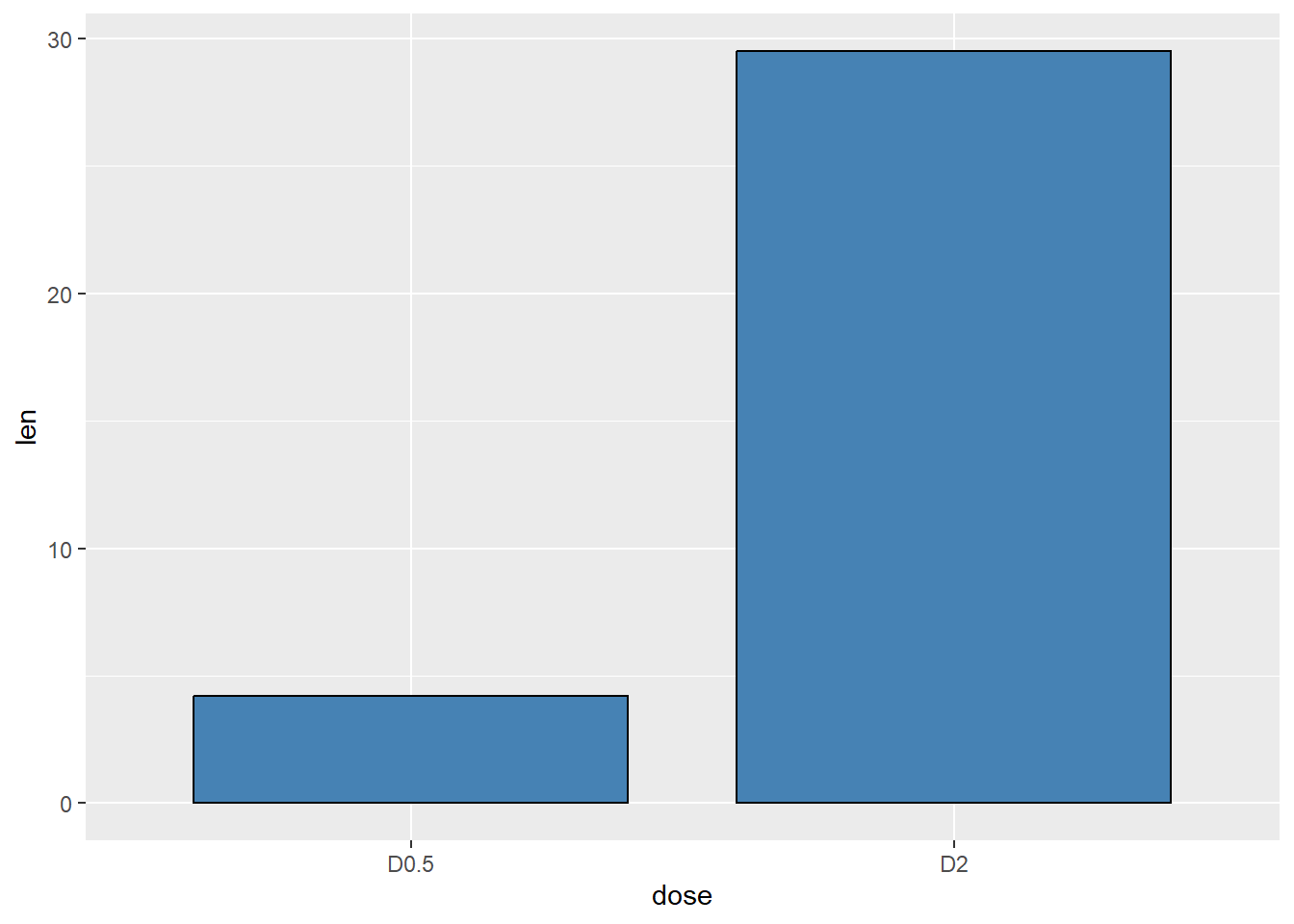
Next we put labels on the bars at the outside and inside
p +
geom_bar(stat = "identity", fill = "steelblue")+
geom_text(aes(label=len), vjust=-0.5, size = 4, col = "black")+
theme_minimal()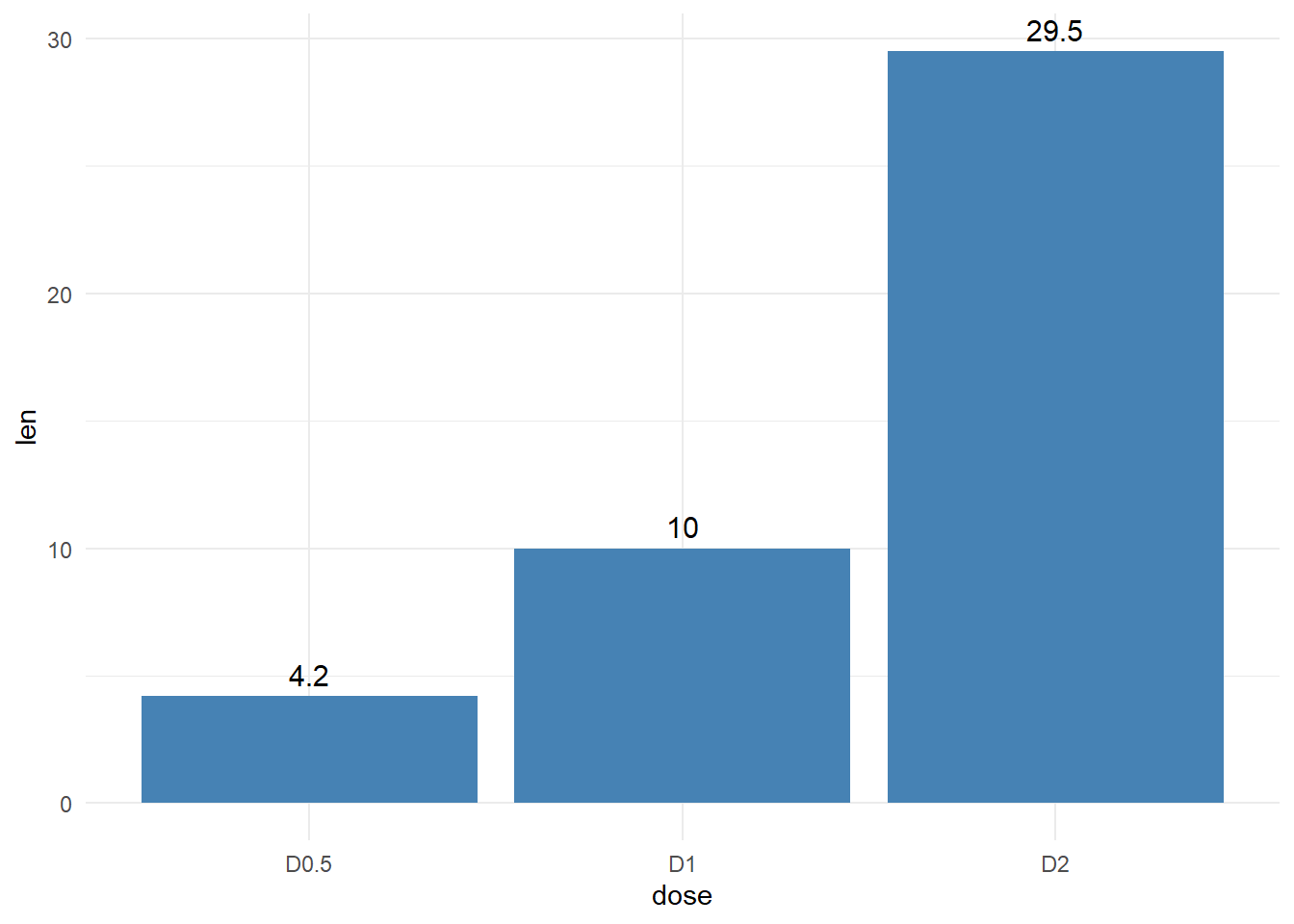
# Change barplot line colors by groups
p <-
ggplot(df, aes(x = dose, y = len, color = dose)) +
geom_bar(stat = "identity", fill = "white") +
geom_text(aes(label=len), vjust=1.5, size=5, col = "red")
p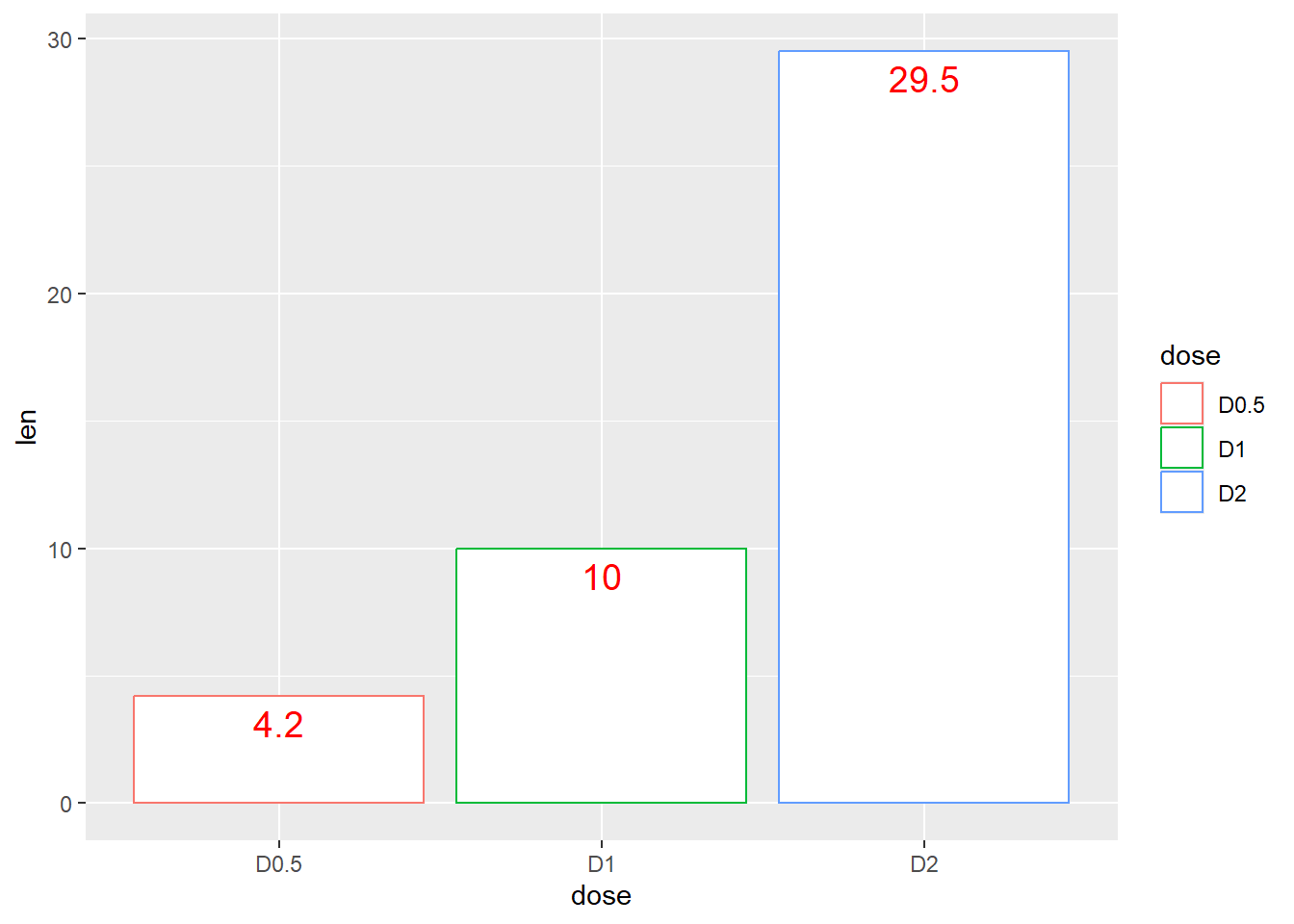
p + scale_color_manual(values=c("#999999", "#E69F00", "#56B4E9"))
Use brewer color palettes
p + scale_color_brewer(palette="Dark2")
p + scale_color_grey() + theme_classic()
Change barplot fill colors by groups
#
p <-
ggplot(df, aes(x=dose, y=len, fill=dose)) +
geom_bar(stat="identity")+theme_minimal()
p
Use custom color palettes
p +
scale_fill_manual(values=c("#999999", "#E69F00", "#56B4E9"))
p +
scale_fill_brewer(palette="Dark2")
Use grey scale
p + scale_fill_grey()
ggplot(df, aes(x=dose, y=len, fill=dose))+
geom_bar(stat="identity", color="black")+
scale_fill_manual(values=c("#999999", "#E69F00", "#56B4E9"))+
theme_minimal()
Change bar fill colors to blues
p +
scale_fill_brewer(palette = "Blues") +
labs(
fill = NULL,
y = "Length (cm)")+
theme(
legend.position = "inside",
legend.position.inside = c(0.2,.8),
legend.direction = "horizontal",
legend.background = element_rect(color = "gray45"))
Change position of bars
p +
scale_x_discrete(limits=c("D2", "D0.5", "D1"))
dat <- foreign::read.dta("C:/dataset/bea_organ_damage_28122013.dta")
BC <-
dat %>%
select(q2idtype, q3sex) %>%
na.omit() %>%
group_by(q2idtype, q3sex) %>%
summarize(Freq = n()) %>%
ggplot(aes(x=q2idtype, y=Freq, fill=q3sex))`summarise()` has grouped output by 'q2idtype'. You can override using the
`.groups` argument.Next we draw the barplot using the economist theme from the ggthemes package
BC +
geom_bar(stat="identity", position= position_dodge()) +
geom_text(aes(label=Freq), vjust=1.6, color="black",
size=4, position = position_dodge(0.9)) +
scale_fill_brewer(palette="Reds") +
labs(title="My Barplot", x="Case or Control", y="Frequency") +
scale_color_discrete(name="Sex") +
ggthemes::theme_stata()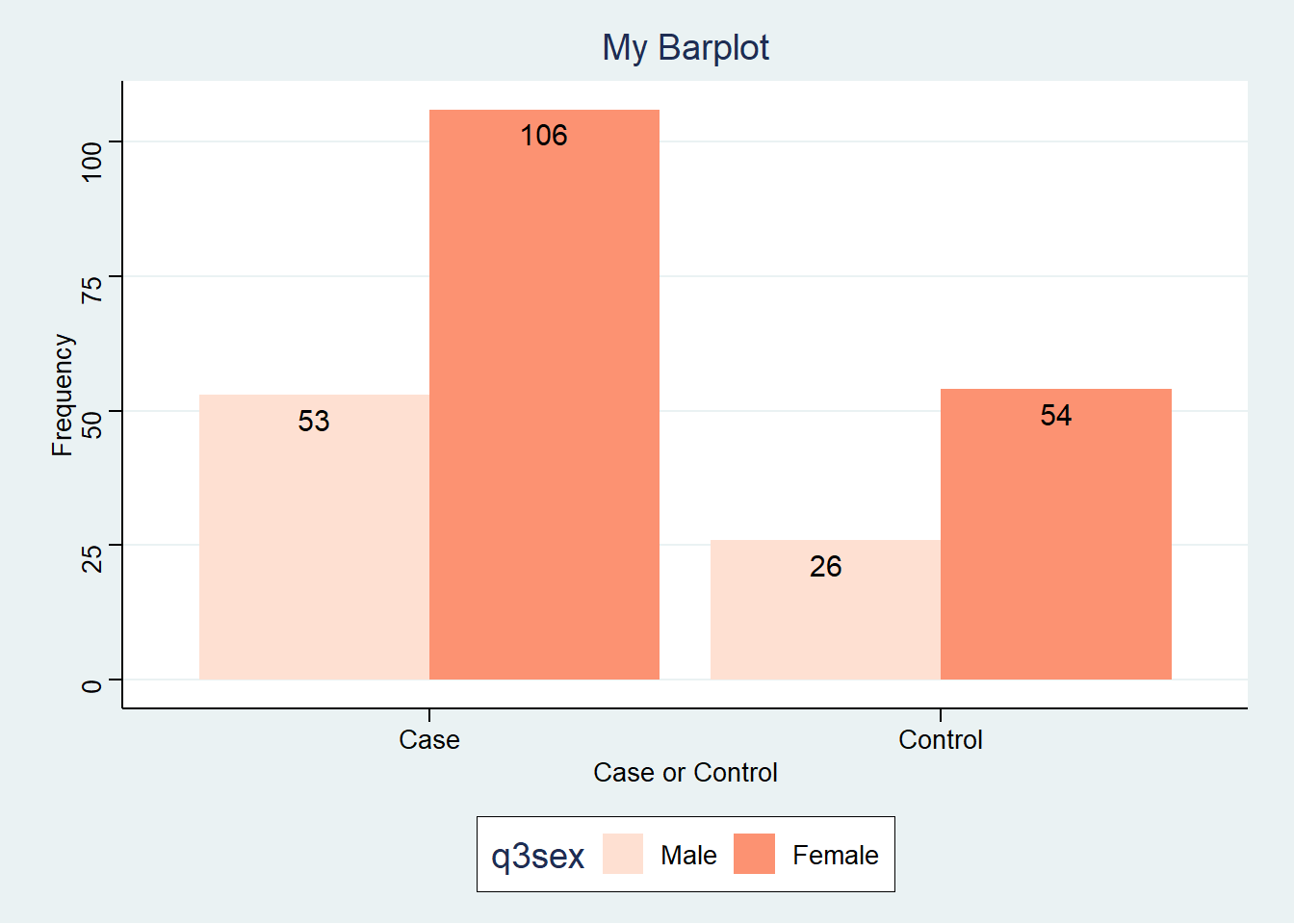
Next we plot a baroplot with error bars. To do that we first we form the ggplot object that we call BC.
BC <-
dat %>%
select(Type = q2idtype, Sex = q3sex, q12weight) %>%
na.omit() %>%
group_by(Type, Sex) %>%
summarize(Mean.wgt = mean(q12weight), SD.wgt = sd(q12weight)) %>%
ggplot(aes(x=Type, y=Mean.wgt, fill=Sex))`summarise()` has grouped output by 'Type'. You can override using the
`.groups` argument.And then plot the graph
BC +
geom_bar(stat="identity", position=position_dodge()) +
geom_errorbar(
aes(ymin = Mean.wgt-SD.wgt, ymax = Mean.wgt+SD.wgt),
width=.2,
size=0.,
position=position_dodge(.9)) +
labs(
title="Mean weight with error bars",
x="Case or Control",
y="Mean(kgs)") +
scale_fill_brewer(palette="Paired") +
ggthemes::theme_stata()Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.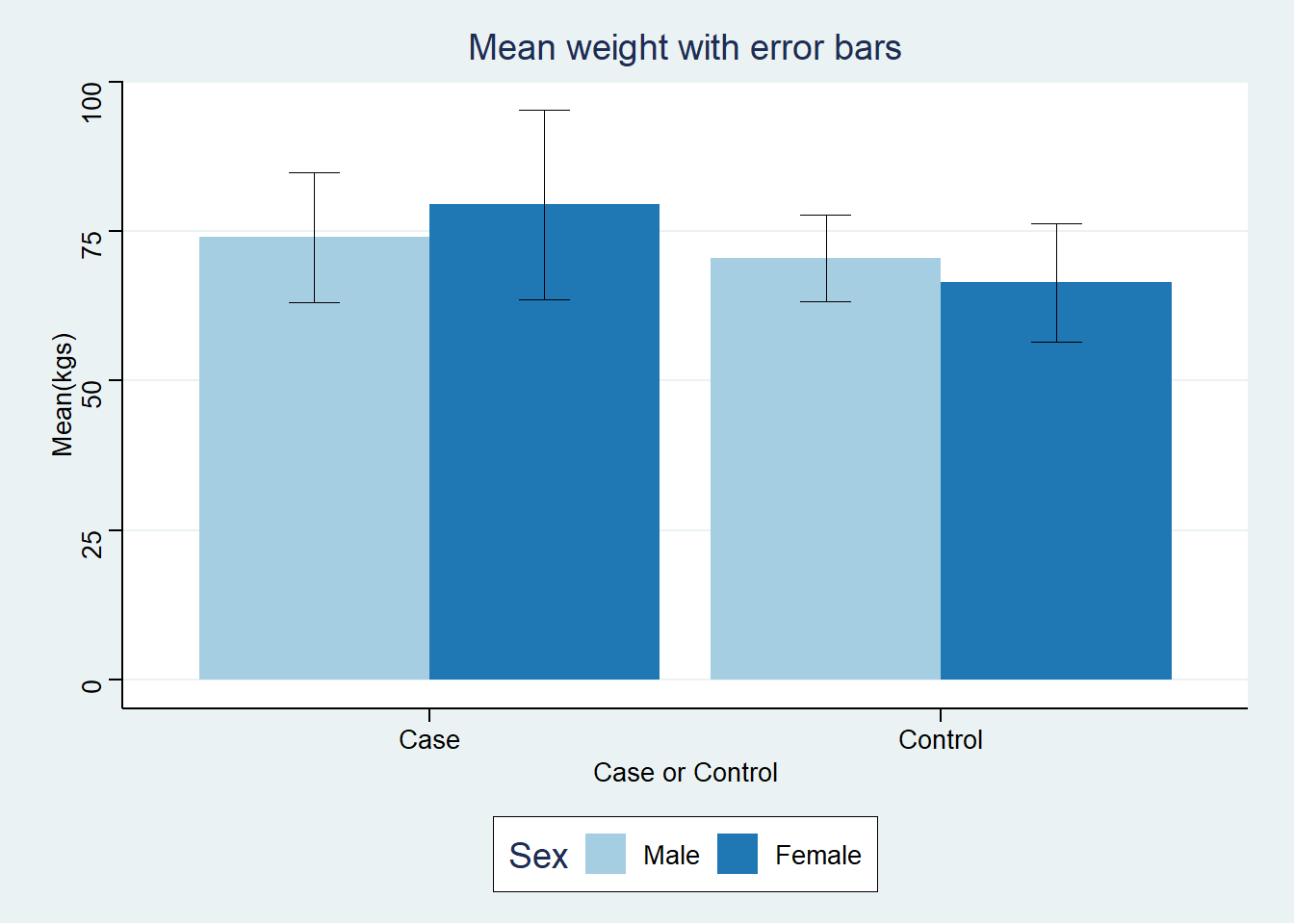
df2 <- data.frame(supp=rep(c("VC", "OJ"), each=3),
dose=rep(c("D0.5", "D1", "D2"),2),
len=c(6.8, 15, 33, 4.2, 10, 29.5))
head(df2)TRUE supp dose len
TRUE 1 VC D0.5 6.8
TRUE 2 VC D1 15.0
TRUE 3 VC D2 33.0
TRUE 4 OJ D0.5 4.2
TRUE 5 OJ D1 10.0
TRUE 6 OJ D2 29.5df2 %>%
ggplot(aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity")
df2 %>%
ggplot(aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity", position=position_dodge())
Change color manually
p <-
ggplot(data=df2, aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity", color="black", position=position_dodge()) +
scale_fill_manual(values=c('#999999','#E69F00')) +
theme_minimal()
p
Create some data
df_sorted <-
tibble(supp = factor(rep(c("VC", "OJ"), each=3)),
dose = rep(c("0.5", "1", "2"),2),
len = c(6.8, 15, 33, 4.2, 10, 29.5)) %>%
arrange(dose, supp) %>%
group_by(dose) %>%
mutate(label_ypos=cumsum(len))
df_sorted# A tibble: 6 × 4
# Groups: dose [3]
supp dose len label_ypos
<fct> <chr> <dbl> <dbl>
1 OJ 0.5 4.2 4.2
2 VC 0.5 6.8 11
3 OJ 1 10 10
4 VC 1 15 25
5 OJ 2 29.5 29.5
6 VC 2 33 62.5df_sorted %>%
ggplot(aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity")+
geom_text(aes(y=label_ypos, label=len), vjust=1.6,
color="white", size=3.5)+
scale_fill_brewer(palette="Paired")+
theme_minimal()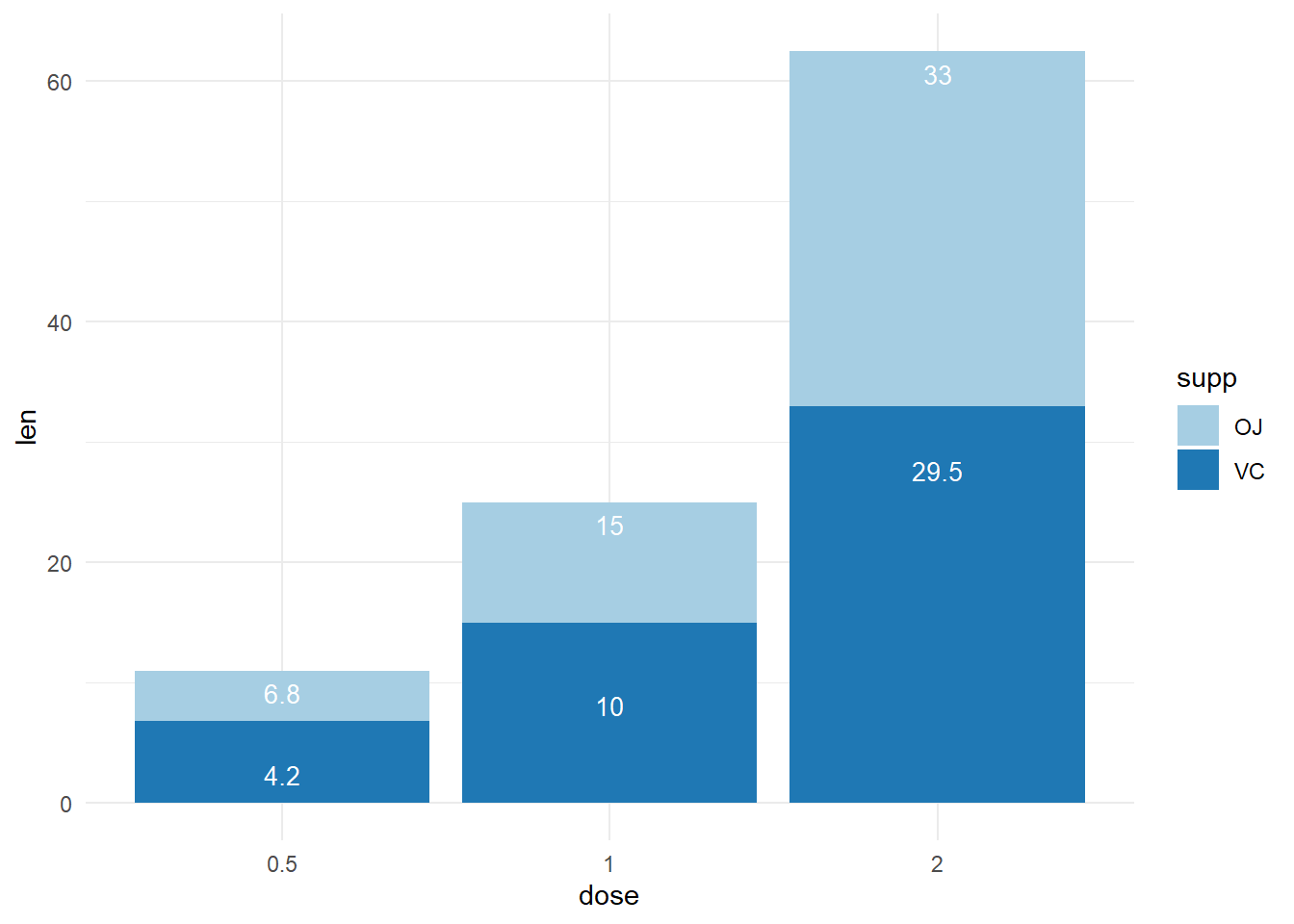
Plotting barplot with x-axis treated as continuous variable
#
df_sorted %>%
mutate(dose = as.numeric(dose)) %>%
ggplot(aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity", position=position_dodge())+
scale_fill_brewer(palette="Paired")+
theme_minimal()
# Axis treated as discrete variable
df_sorted %>%
mutate(dose = as.factor(dose)) %>%
ggplot(aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity", position=position_dodge())+
scale_fill_brewer(palette="Paired")+
theme_minimal()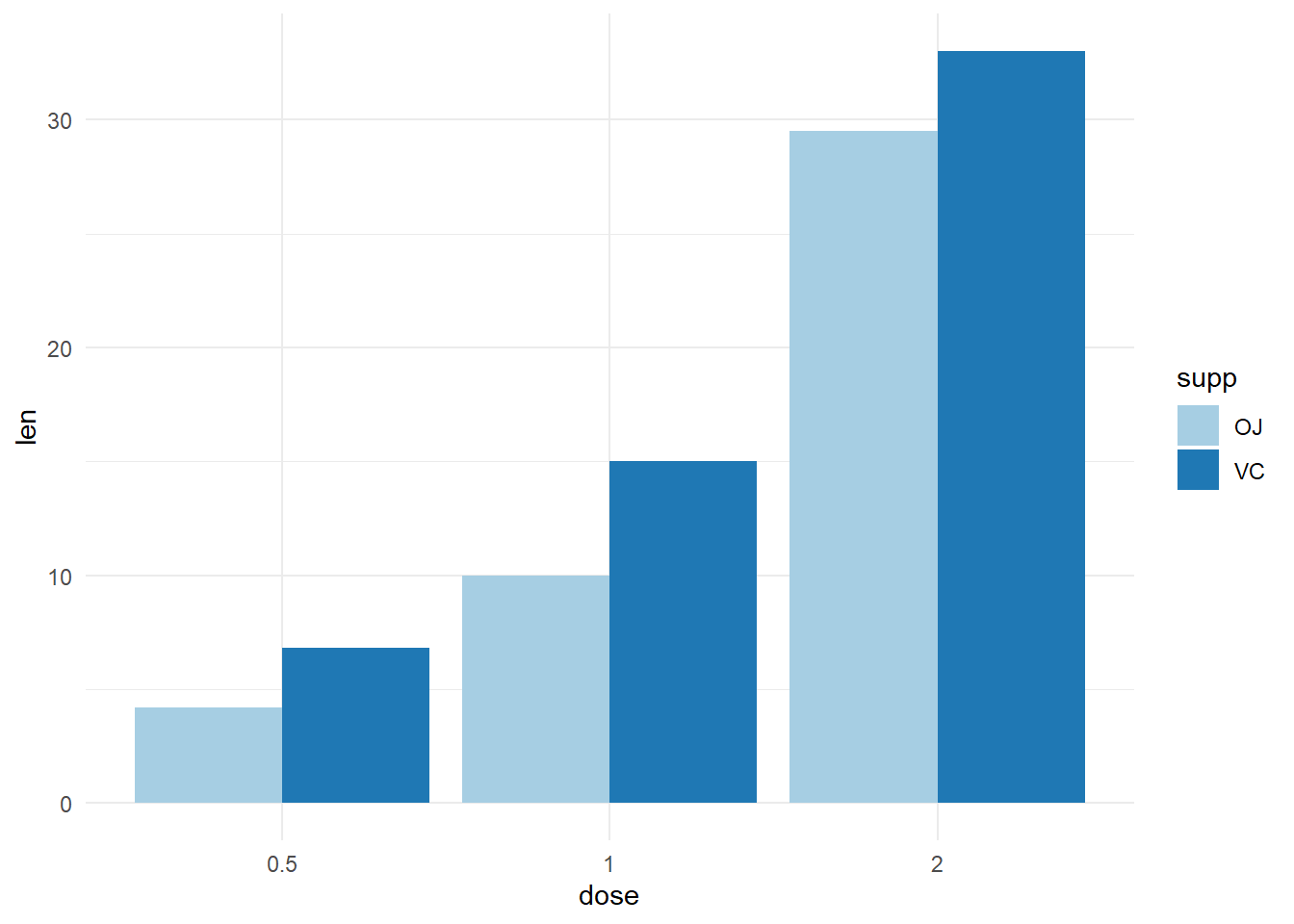
ToothGrowth %>%
mutate(dose = as.factor(dose)) %>%
group_by(supp, dose) %>%
summarise(sd = sd(len), len = mean(len), .groups = "drop") %>%
ggplot(aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=len-sd, ymax=len+sd), width=.2,
position=position_dodge(.9)) +
labs(title="Plot of length per dose", x="Dose (mg)", y = "Length")+
scale_fill_brewer(palette="Paired") +
theme_minimal()
babies <-
dget("babies_clean") %>%
select(apgar5cat, died)
babies %>%
ggplot(aes(x = apgar5cat, fill = apgar5cat)) +
geom_bar(stat = 'count') +
scale_fill_grey(
name = "APGAR Category",
label = c("LOW", "MEDIUM", "HIGH"))+
theme_bw()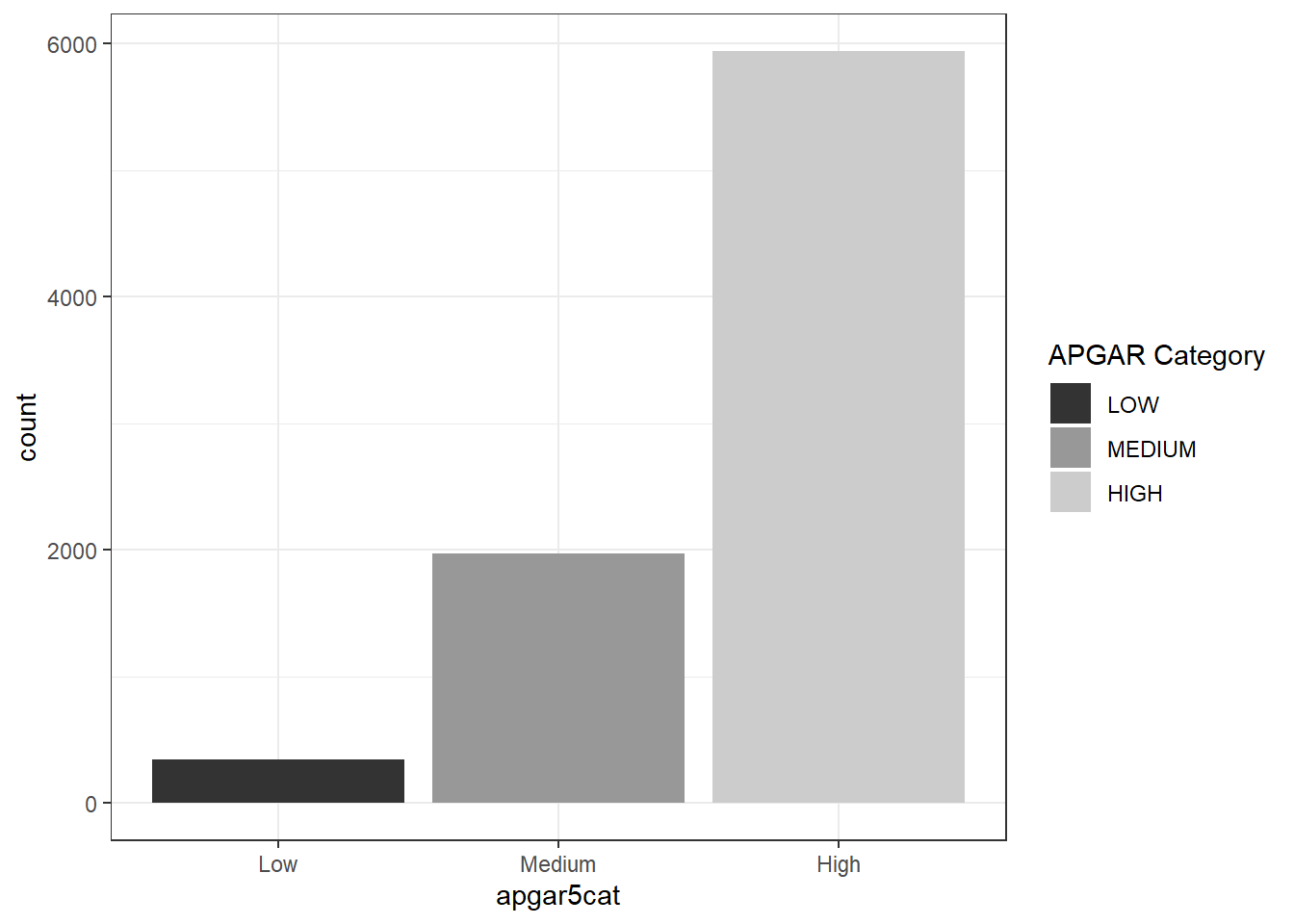
Graph of binomial confiedence intervals
tibble(
alg = c(
"D farina", "D pteronyssinus", "American Cockroach",
"Dog","Alternaria","Rye grass"),
n = c(125, 118, 62, 3, 3, 0)) %>%
mutate(N = rep(161, 6)) %>%
mutate(xx = epiDisplay::ci.binomial(n, N)) %>%
unnest(xx) %>%
ggplot(
aes(
x = alg, y = probability, color = alg,
ymin = exact.lower95ci, ymax = exact.upper95ci)) +
geom_col(
fill = "steelblue",
color = "steelblue", alpha = 0.4) +
geom_errorbar(color = "steelblue", width = 0.15)+
labs(
y = "Percentage (%)",
x = NULL,
caption = "Asthma study (2023)")+
theme_bw()+
scale_y_continuous(br = seq(0, 1, 0.2), labels = seq(0, 100, 20))+
theme(
axis.text.x = element_text(
angle = 90, hjust = 1, vjust = 0.05,
face = "italic"))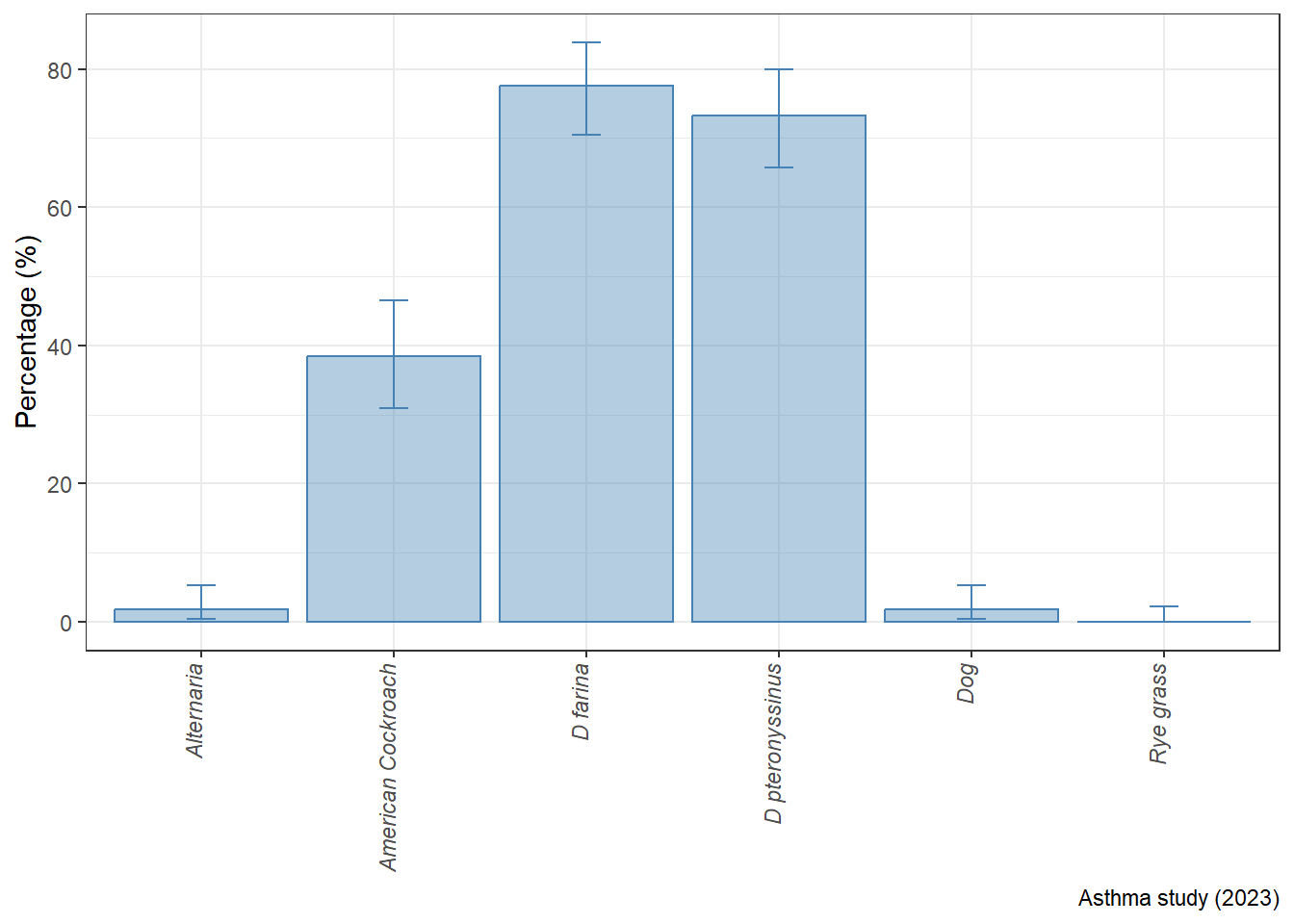
dataF %>%
mutate(
avehb_cat = case_when(
avehb < median(avehb) ~ "Low HB",
avehb >= median(avehb) ~ "High HB") %>%
factor(levels = c("Low HB", "High HB"))) %>%
select(starts_with("mcv"), avehb_cat) %>%
pivot_longer(
cols = c(mcv1:mcv5),
values_to = "mcv",
names_to = "measure") %>%
group_by(avehb_cat, measure) %>%
mutate(
mean_mcv = mean(mcv),
low_mcv = mean_mcv - 1.96*sd(mcv)/sqrt(n()),
high_mcv = mean_mcv + 1.96*sd(mcv)/sqrt(n())) %>%
ggplot(aes(y = mean_mcv, x = measure, fill = avehb_cat)) +
geom_bar(stat = "identity", position = position_dodge(0.9)) +
geom_errorbar(
aes(ymin = low_mcv, ymax = high_mcv), size = 0.8,
width = 0.3, position = position_dodge(0.9)) +
labs(y = "Mean Corpuscular Volume",
x = NULL,
title = "Variation in MCV per review period (95%CI)",
caption = "Source: Data One")+
scale_fill_manual(name = "Hemoglobin \n Status", values = c("grey", "grey45"))+
scale_x_discrete(
labels = toupper(c("First", "Second", "Third", "Fourth", "Fifth")))+
scale_y_continuous(limits = c(0,130), breaks = seq(0, 130, 10))+
theme(
panel.background = element_rect(colour = "black", fill = "white"),
plot.background = element_rect(fill = "grey"),
plot.title = element_text(
face = "bold", hjust = 0.5, family = "serif"),
axis.text = element_text(face = "bold", family = "serif"),
axis.title = element_text(family = "serif", face = "bold"),
plot.caption = element_text(family = "serif", face = "bold"),
legend.text = element_text(family = "serif", face = "bold"),
legend.title = element_text(
family = "serif", face = "bold", hjust = 0))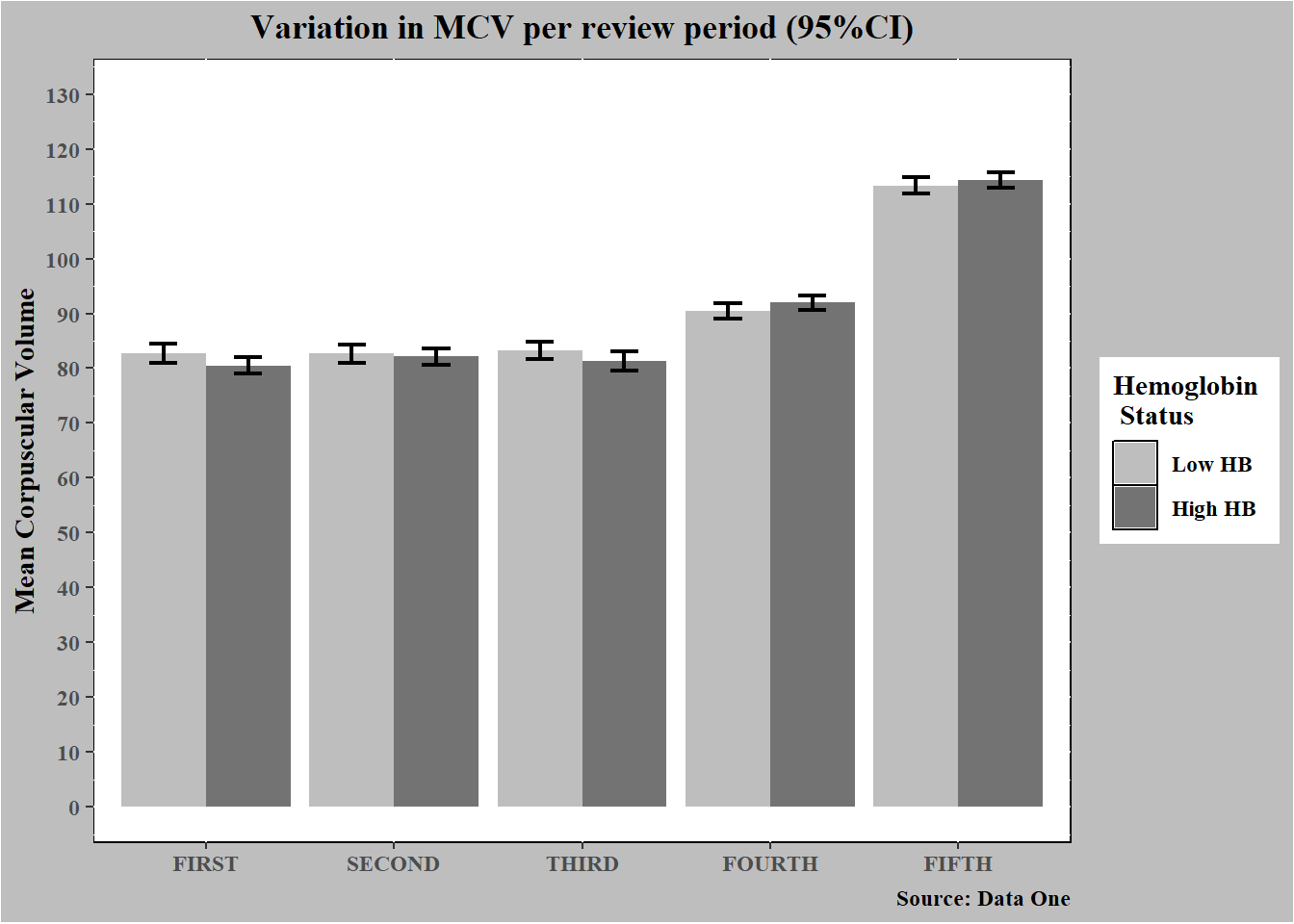
dataF %>%
group_by(anemia1, agecat) %>%
mutate(
Anemia.1 = case_when(
anemia1 == 0 ~ "No",
anemia1 == 1 ~ "Yes") %>% as_factor()) %>%
reframe(
across(
c(hb1,hb2),
list(Mean = mean, SD = sd, se = ~sd(.x)/sqrt(n())))) %>%
ggplot(aes(x = anemia1, y=hb1_Mean, fill = agecat)) +
geom_errorbar(aes(ymin = hb1_Mean - 1.96*hb1_SD, ymax = hb1_Mean + 1.96*hb1_SD),
position = position_dodge(0.9), width = 0.2, size = 0.8) +
geom_bar(
stat = "identity", position = position_dodge(0.9), col = "black") +
labs(
x = "First Anemia Present", y = "Mean of First HgB (mg/dL)",
title = "Average initial HgB for first anemia and Age Categories") +
theme_bw()+
scale_fill_brewer(name = "Age Group", palette = "Dark2",
labels= c("10-19 yrs", "20-29 yrs", "30-39 yrs", "40-49 yrs"))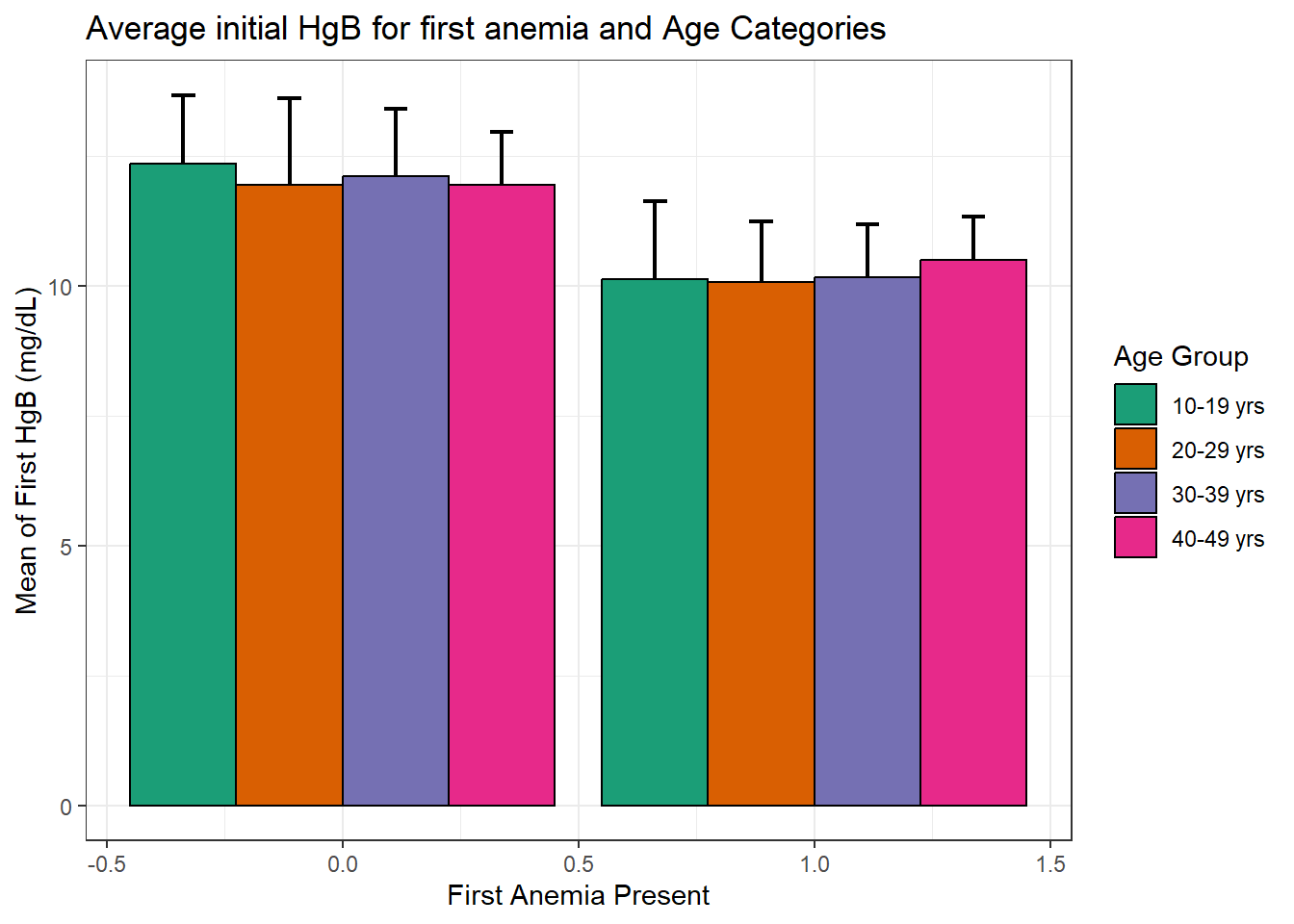
# df_sbp <-
readxl::read_xlsx("C:/Dataset/SBPDATA.xlsx") %>%
janitor::clean_names() %>%
drop_na() %>%
select(disease_class, sbp_0:sbp_18) %>%
pivot_longer(cols = sbp_0:sbp_18) %>%
group_by(disease_class, name) %>%
summarize(across(value, ~epiDisplay::ci(.)), .groups = "drop") %>%
unnest(cols = c(value)) %>%
mutate(
name = factor(
name,
levels = c(
"sbp_0", "sbp_2", "sbp_4", "sbp_6", "sbp_8",
"sbp_10", "sbp_12", "sbp_14", "sbp_16", "sbp_18"),
labels = c(
"Month Zero", "Month Two", "Month Four",
"Month Six", "Month Eight", "Month Ten",
"Month Twelve", "Month Fourten",
"Month Sixteen", "Month Eighteen" )),
disease_class = factor(
disease_class, levels = c("DM", "HPT", "DM+HPT"))) %>%
ggplot(aes(x = disease_class, y = mean, fill = name), col = "black") +
geom_col(position = position_dodge(0.9), col = "gray45", alpha = 0.4)+
geom_errorbar(aes(ymax = mean+se, ymin = mean-se),position = position_dodge(0.9), width = 0.3)+
labs(fill = NULL, x = NULL, y = "Systolic Blood Pressure (mmHg)")+
scale_fill_manual(
values = c("#FF5733","#33FF57","#3357FF","#FFD700","#7D33FF",
"#FF33C1", "#33FFF5","#FF8C33","#8CFF33",
"#F533FF"))+
scale_y_continuous(limits = c(0, 150))+
scale_x_discrete(
limits = c("DM", "HPT", "DM+HPT"),
label = c(
"Diabetes Mellitus",
"Hypertension",
"Diabetes mellitus and \n Hypertension"))+
coord_cartesian(ylim = c(100, 150), expand = F)+
theme_bw()+
theme(
legend.position = "bottom",
axis.title.y = element_text(face = "bold"),
axis.text.x = element_text(face = "bold", size = 11))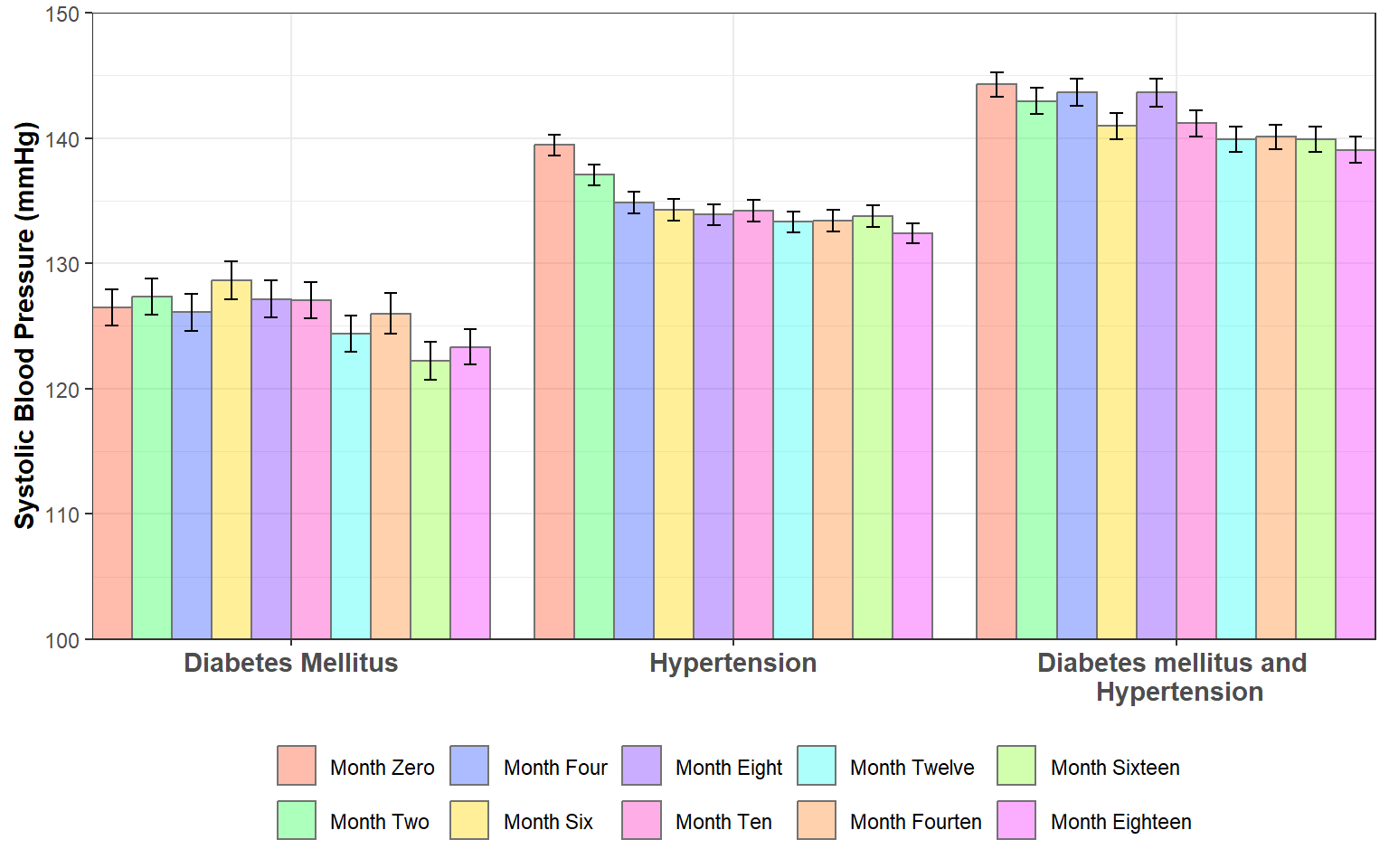
library(showtext)
library(ggtext)
font_add_google("Libre Franklin", "franklin")
font_add_google("Gelasio", "gelasio")
showtext_opts(dpi = 300)
showtext_auto()
data <- tribble(
~category,~gain,~none,~lose,~group,
"Total",21,37,41,"total",
"Men",24,42,34,"gender",
"Women",18,34,47,"gender",
"White",24,43,33,"race",
"Black",11,20,66,"race",
"Hispanic",17,29,52,"race",
"Asian*",15,38,47,"race",
"Ages 18-29",22,30,47,"age",
"30-49",22,34,42,"age",
"50-64",21,42,37,"age",
"65+",17,43,39,"age",
"Postgrad",17,38,45,"education",
"College grad",17,43,39,"education",
"Some college",22,33,43,"education",
"HS or less",23,37,40,"education",
"Rep/Lean Rep",35,51,13,"politics",
"<span style='color:gray50'>Conserv</span>",39,54,6,"politics",
"<span style='color:gray50'>Mod/Lib</span>",29,47,23,"politics",
"Dem/Lean Dem",7,23,69,"politics",
"<span style='color:gray50'>Cons/Mod</span>",8,25,66,"politics",
"<span style='color:gray50'>Liberal</span>",6,20,74,"politics"
) %>%
mutate(group = factor(
group,
levels = c("total", "gender", "race","age", "education", "politics")),
category = factor(category, levels = rev(category)))
data %>%
pivot_longer(
-c(category, group),
names_to = "effect",
values_to = "percent") %>%
mutate(
effect = factor(effect, levels = c("lose", "none", "gain"))) %>%
ggplot(aes(x = percent, y = category, fill = effect, label = percent)) +
geom_col(width = 0.7) +
geom_text(
position = position_stack(vjust = 0.5),
family = "franklin", size = 6, size.unit = "pt") +
facet_grid(group~., scales = "free_y", space = "free_y") +
coord_cartesian(expand = FALSE, clip = "off") +
scale_fill_manual(
name = NULL,
breaks = c("gain", "none", "lose"),
values = c("#D0A62D", "#DBD9C6", "#E3CA83"),
labels = c("Gain influence", "Not be affected", "Lose influence")) +
labs(
title = "How Americans feel people like them<br>will fare under the Trump administration",
subtitle = "*% who say **people like yourself** will in<br>Washington with Donald Trump taking office*",
caption = "\\* Estimates for Asian adults are representative of English speakers<br>only.<br>Note: No answer responses are not shown.<br>Source: Survey of U.S. adults conducted Jan. 27-Feb. 2, 2025.<br><br>**<span style='color:black'>PEW RESEARCH CENTER</span>**",
x = NULL,
y = NULL) +
theme(
text = element_text(family = "franklin"),
legend.position = "top",
legend.justification.top = "right",
legend.text = element_text(size = 6, margin = margin(l = 2)),
legend.key.size = unit(5, "pt"),
legend.key.spacing.x = unit(2, "pt"),
legend.box.spacing = unit(8, "pt"),
legend.margin = margin(r = 0),
panel.grid = element_blank(),
panel.background = element_blank(),
strip.text = element_blank(),
axis.ticks = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_markdown(size = 6, color = "black"),
plot.title.position = "plot",
plot.title = element_markdown(
face = "bold", size = 9, lineheight = 1.4),
plot.subtitle = element_markdown(
family = "gelasio", size = 8,
color = "gray40", lineheight = 1.2,
margin = margin(b = 8)),
plot.caption.position = "plot",
plot.caption = element_markdown(
hjust = 0, size = 5.5, lineheight = 1.4,
color = "gray40", margin = margin(t = 16)),
plot.margin = margin(t = 10, r = 8, b = 10, l = 3),
panel.spacing.y = unit(15, "pt"))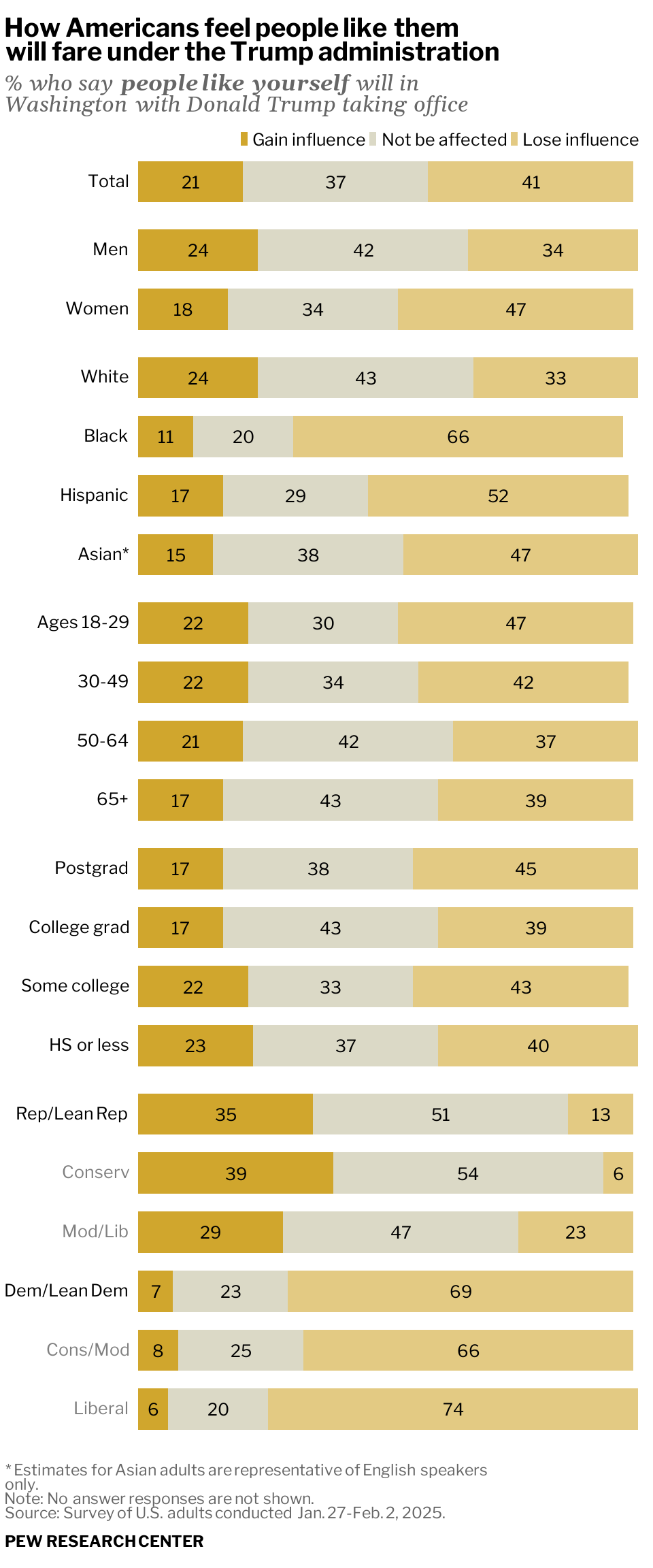
ggsave("gainers_losers.png", width = 2.53, height = 6)