Read in Data
We begin by importing the data into R Studio and then summarizing it.
Code
<- :: read.dta13 ("C:/Dataset/olivia_data_wide.dta" ) %>% select (hb1, hb2, hb3, hb4)%>% :: dfSummary (labels.col = F, graph.col = F)
Data Frame Summary
df_histo
Dimensions: 350 x 4
Duplicates: 3
-----------------------------------------------------------------------------------
No Variable Stats / Values Freqs (% of Valid) Valid Missing
---- ----------- ------------------------ -------------------- ---------- ---------
1 hb1 Mean (sd) : 11.3 (1.2) 57 distinct values 350 0
[numeric] min < med < max: (100.0%) (0.0%)
8.3 < 11.3 < 16.6
IQR (CV) : 1.8 (0.1)
2 hb2 Mean (sd) : 11.2 (1.3) 63 distinct values 350 0
[numeric] min < med < max: (100.0%) (0.0%)
6.1 < 11 < 15.6
IQR (CV) : 1.8 (0.1)
3 hb3 Mean (sd) : 11.1 (1.2) 57 distinct values 350 0
[numeric] min < med < max: (100.0%) (0.0%)
8 < 11.1 < 15.2
IQR (CV) : 1.8 (0.1)
4 hb4 Mean (sd) : 11.8 (2.5) 89 distinct values 350 0
[numeric] min < med < max: (100.0%) (0.0%)
3.5 < 11.5 < 24.4
IQR (CV) : 2.4 (0.2)
-----------------------------------------------------------------------------------
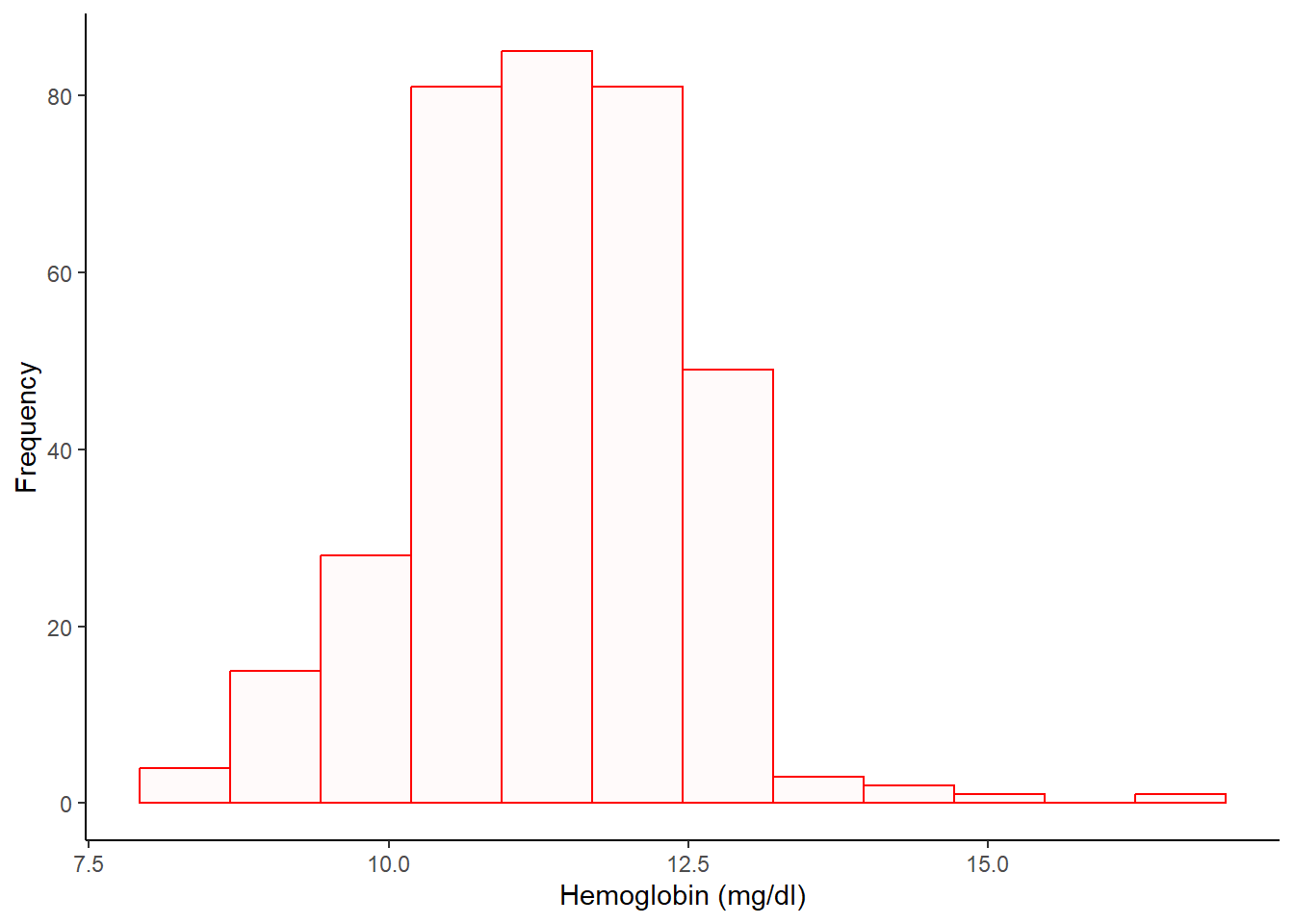
Simple histogram
Code
%>% ggplot (aes (x = hb1)) + geom_histogram (col = "red" , fill = "snow1" , bins = 12 ) + labs (x = "Hemoglobin (mg/dl)" , y = "Frequency" ) + theme_classic ()
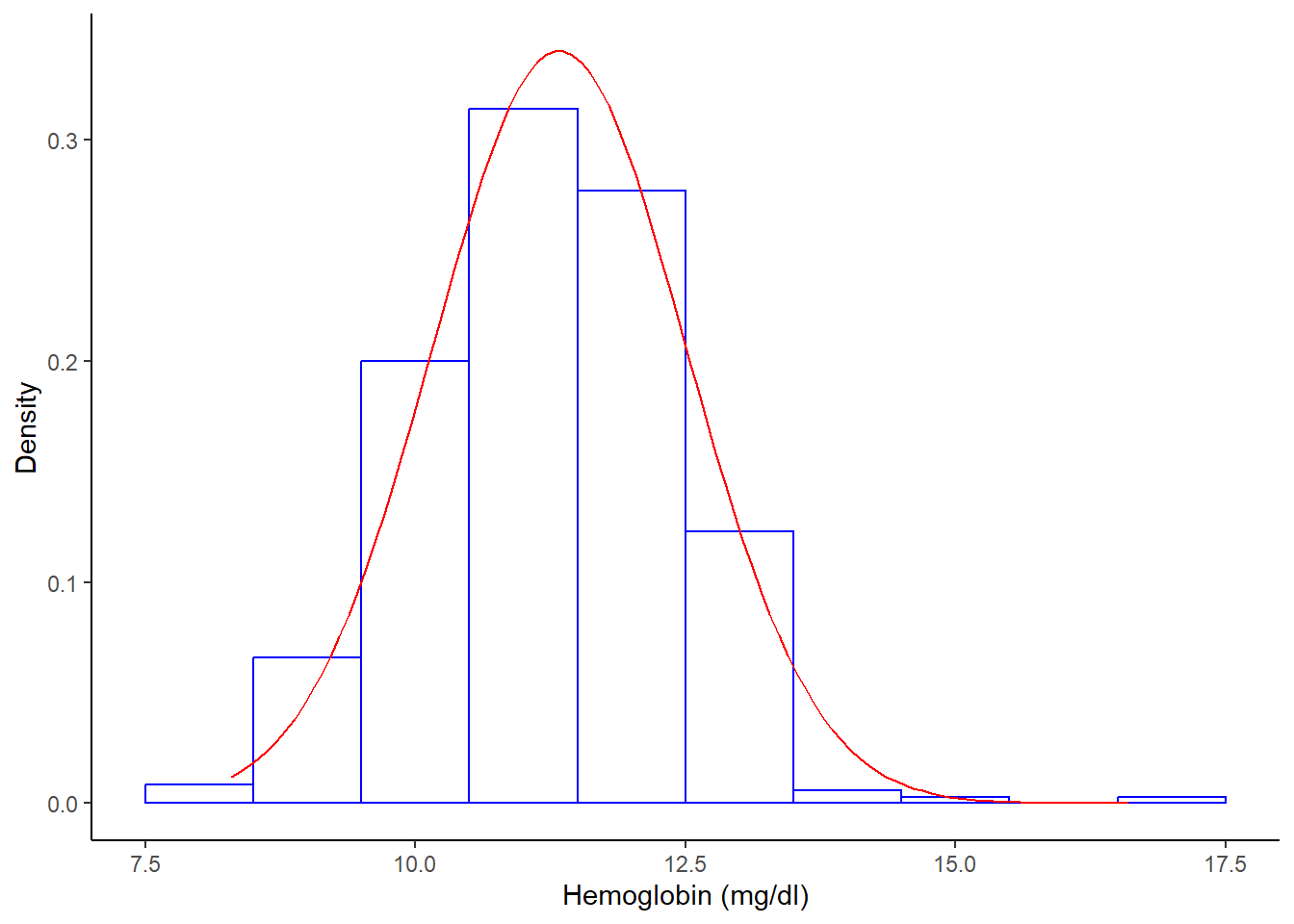
Histogram with normal curve
Code
%>% ggplot (aes (x = hb1)) + geom_histogram (aes (y = after_stat (density)),breaks = seq (7.5 , 17.5 , by = 1 ), colour = "blue" , fill = "white" ) + stat_function (fun = dnorm, args = list (mean = mean (df_histo$ hb1), sd = sd (df_histo$ hb1)),color = 'red' )+ labs (x = "Hemoglobin (mg/dl)" , y = "Density" ) + theme_classic ()
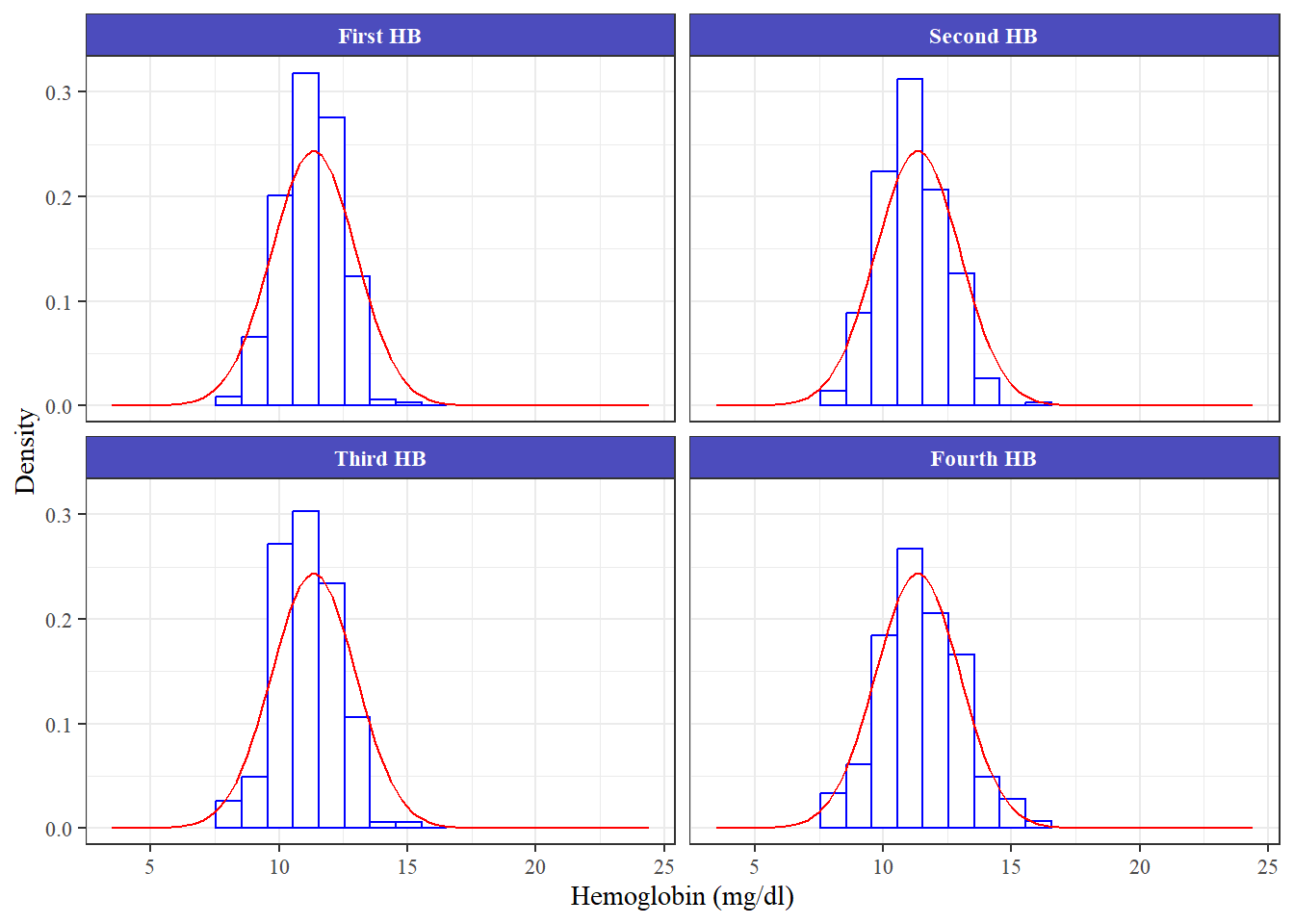
Panel histogram
Code
<- %>% pivot_longer (cols = c (hb1, hb2, hb3, hb4)) %>% drop_na (value) %>% mutate (name = factor (levels = c ("hb1" , "hb2" , "hb3" , "hb4" ),labels = c ("First HB" , "Second HB" , "Third HB" , "Fourth HB" )))%>% ggplot (aes (x = value)) + geom_histogram (aes (y = after_stat (density)),breaks = seq (7.55 , 17.5 , by = 1 ), colour = "blue" , fill = "white" , bins = 10 ) + stat_function (fun = dnorm, args = list (mean = mean (df_temp$ value), sd = sd (df_temp$ value)),color = 'red' )+ labs (x = "Hemoglobin (mg/dl)" , y = "Density" ) + theme_bw ()+ facet_wrap (facets = .~ name)+ theme (text = element_text (family = "serif" ),strip.text = element_text (face = "bold" , color = "white" ),strip.background = element_rect (fill = "#4C4CBD" ),plot.title = element_text (face = 'bold' ))
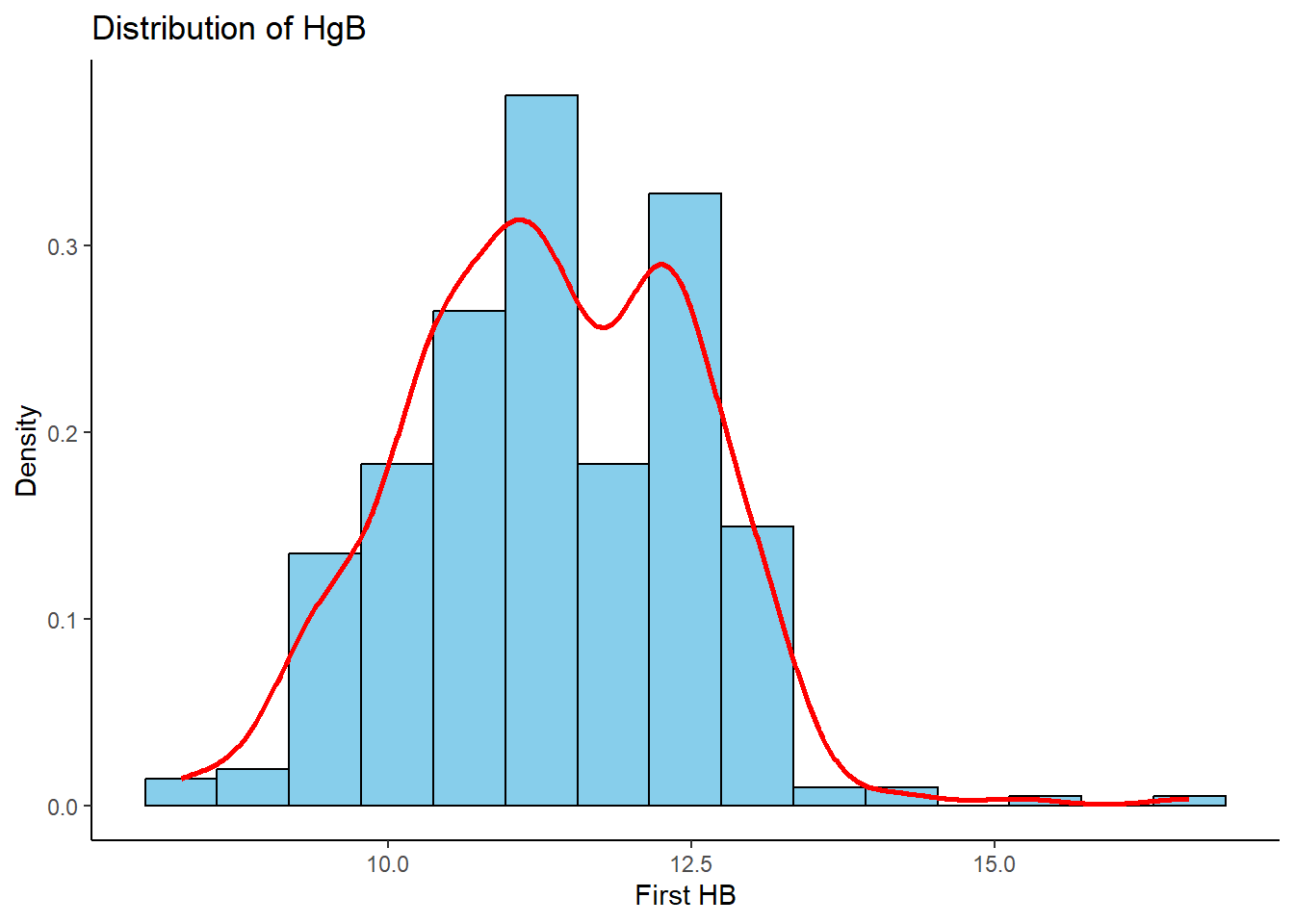
Histogram with density overlay
Code
%>% ggplot (aes (x = hb1, y = ..density..)) + geom_histogram (fill = "skyblue" , col = "black" , bins = 15 )+ geom_density (aes (y = ..density..), col = "red" , size= 1 ) + labs (x = "First HB" , y = "Density" , title = "Distribution of HgB" )+ theme_classic ()
Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
ℹ Please use `after_stat(density)` instead.