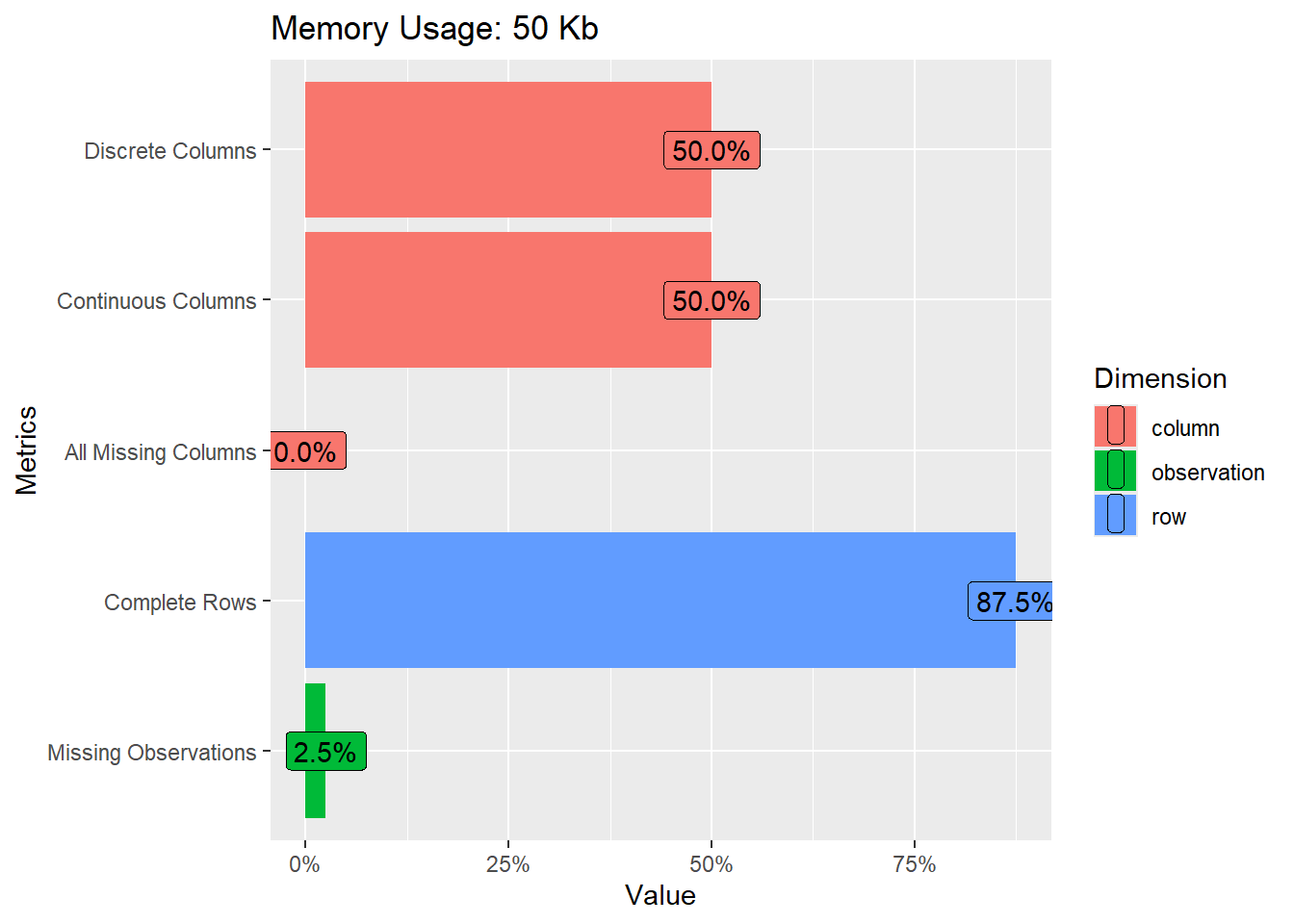
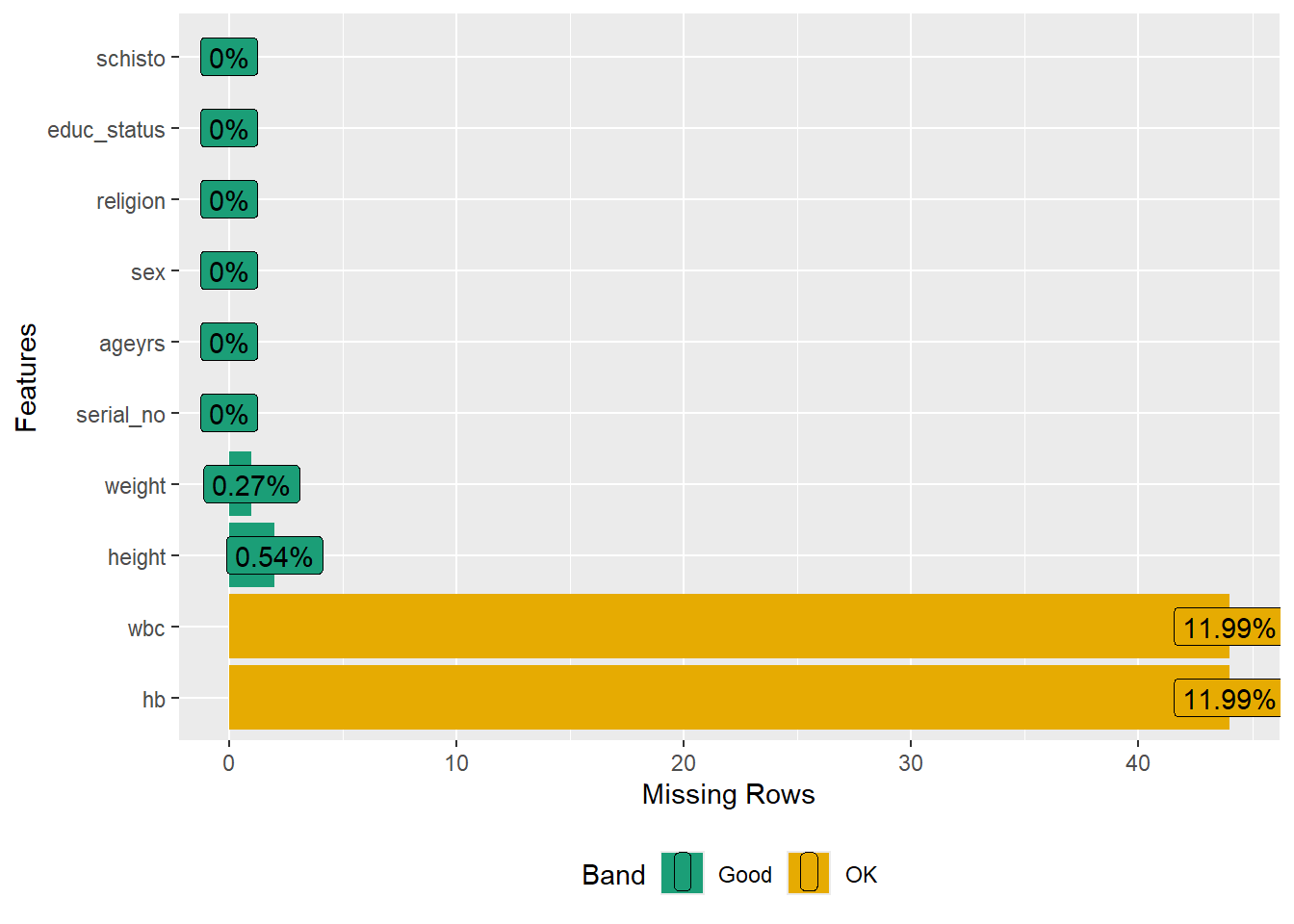
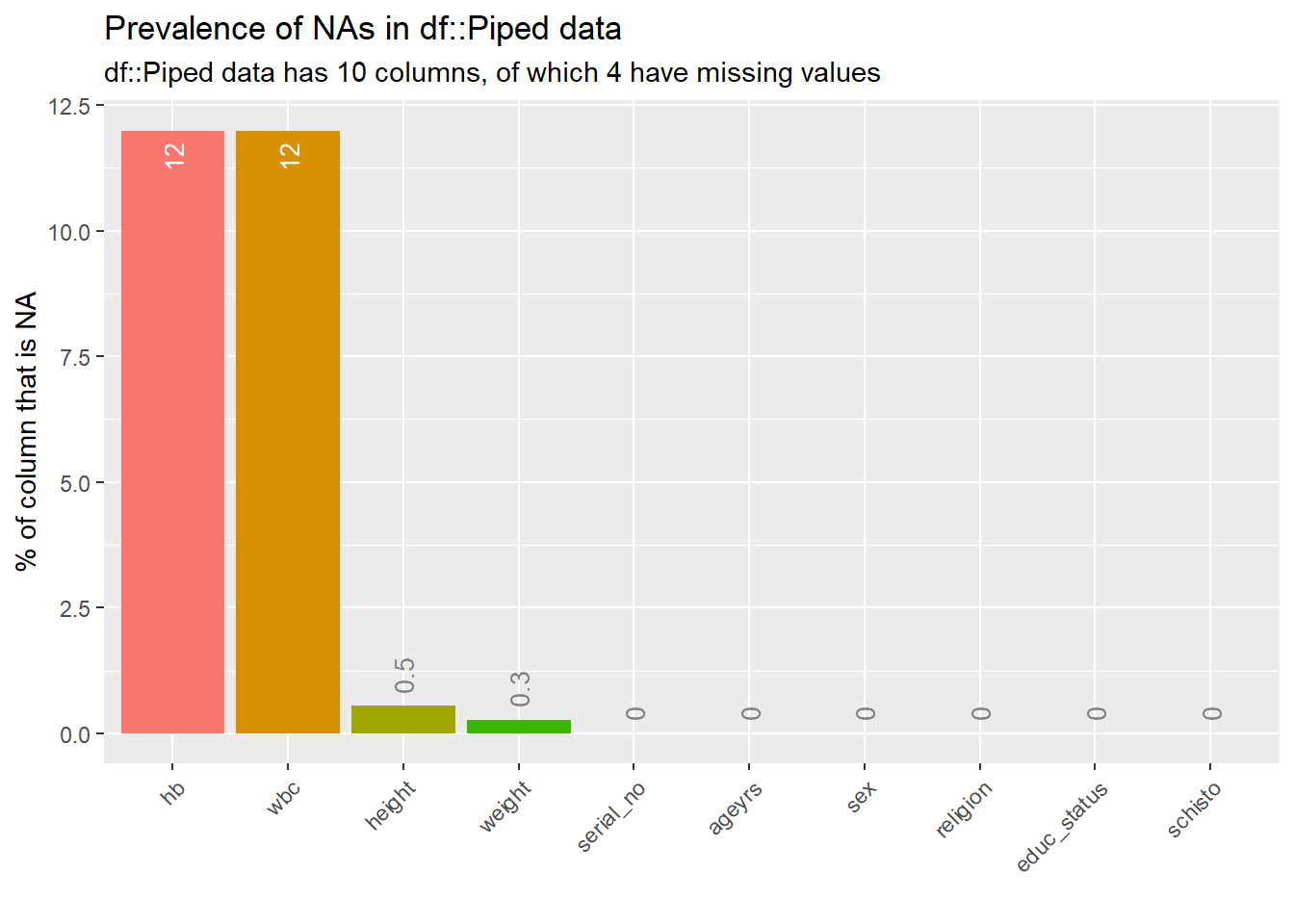
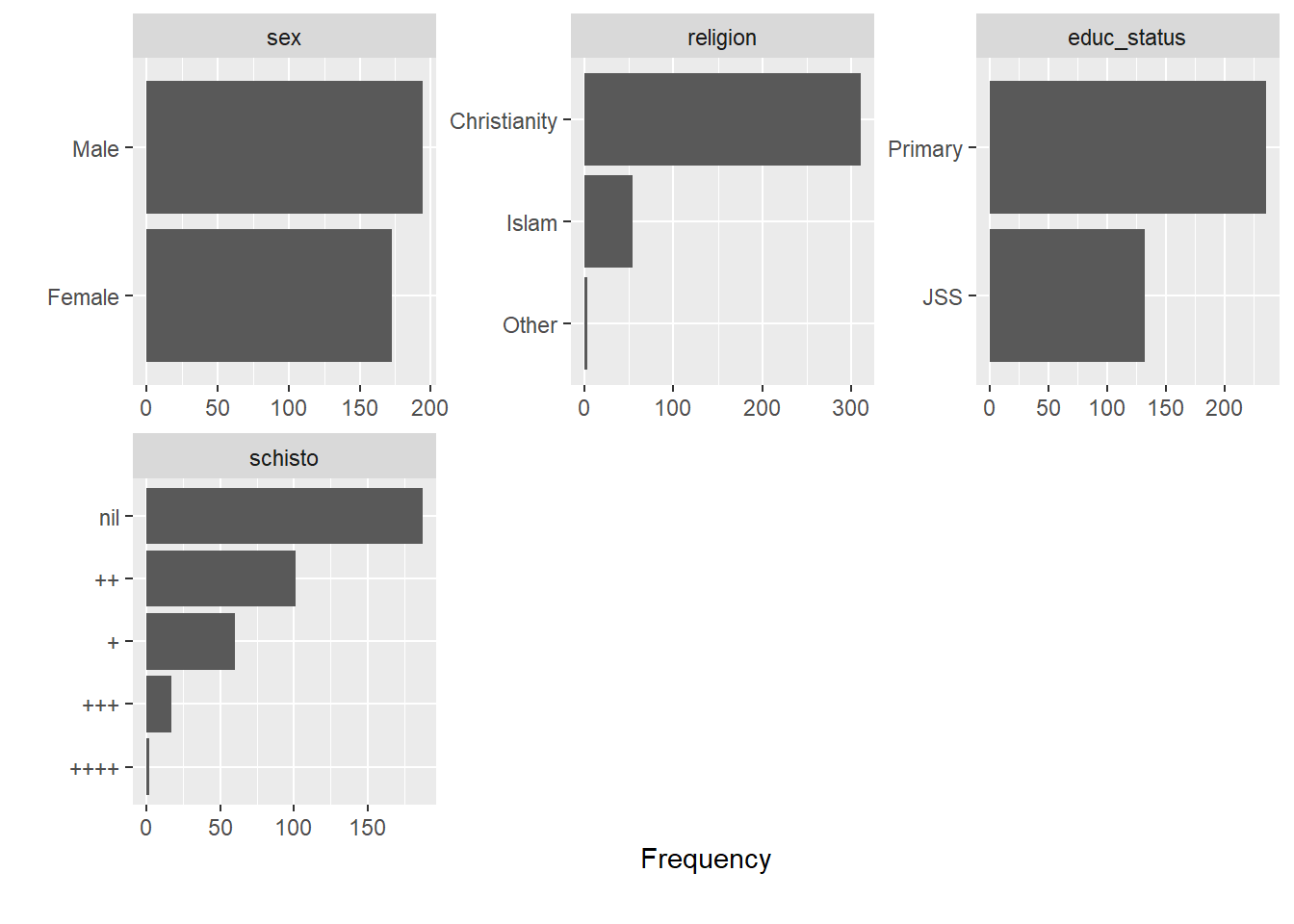
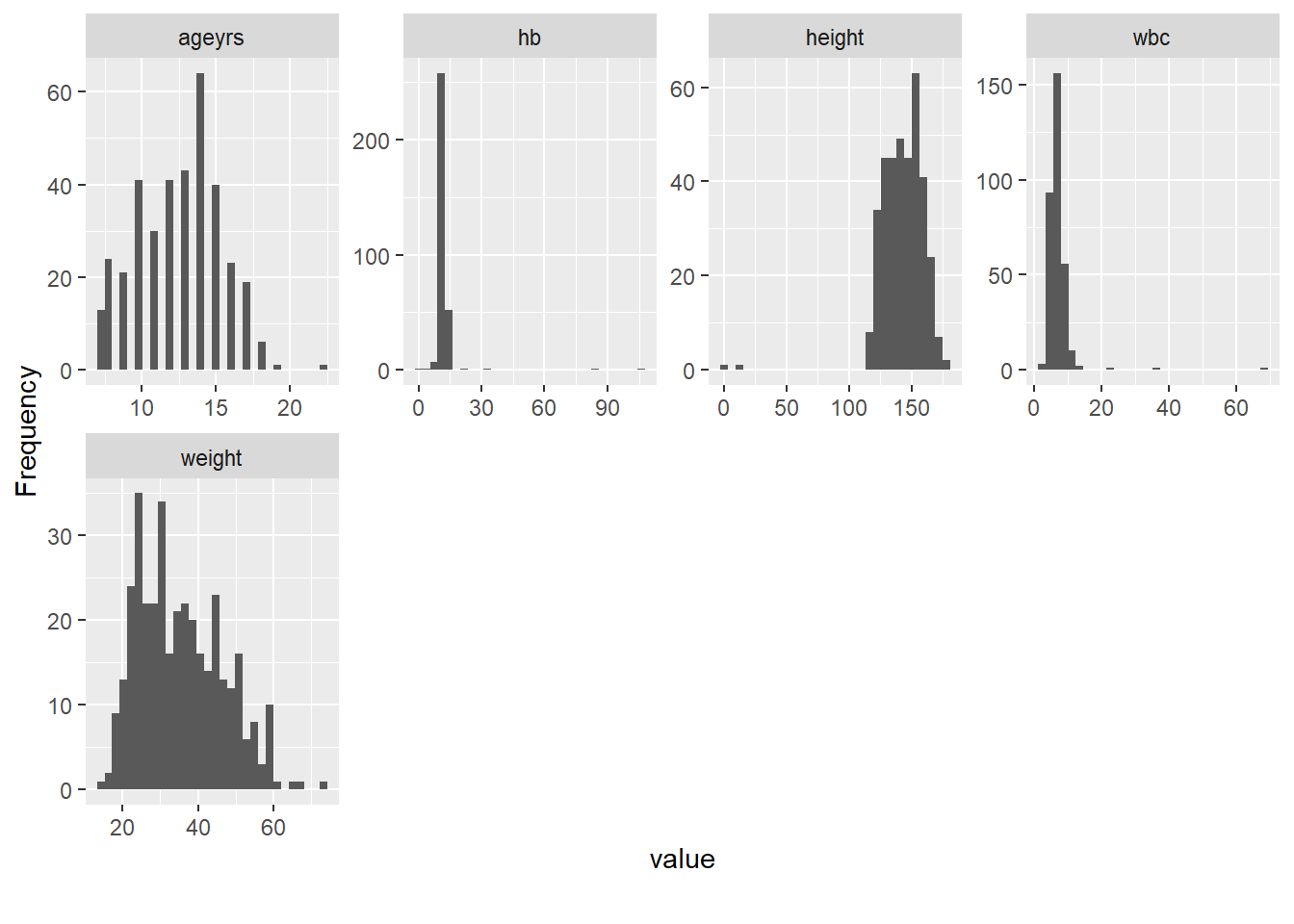
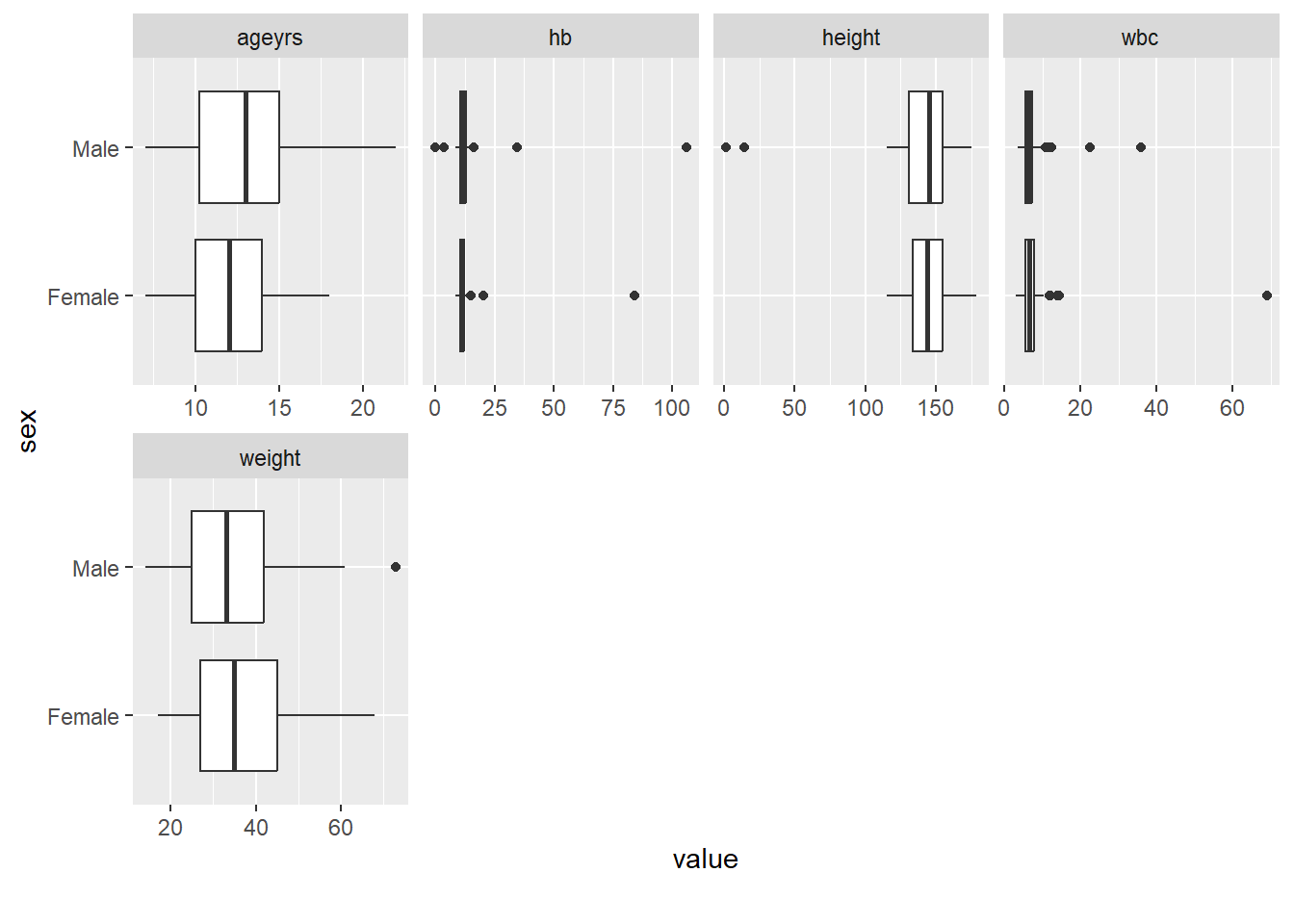
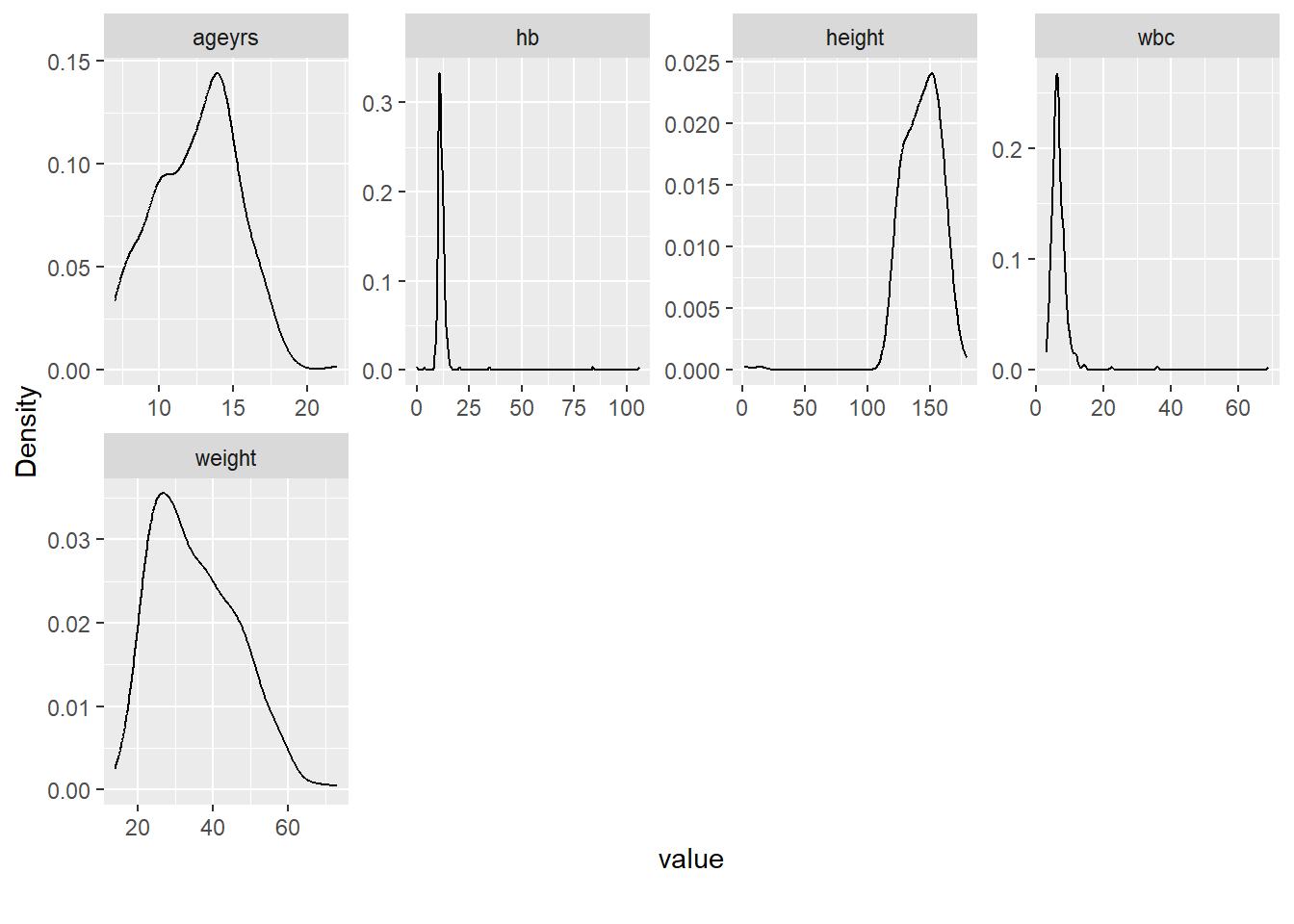
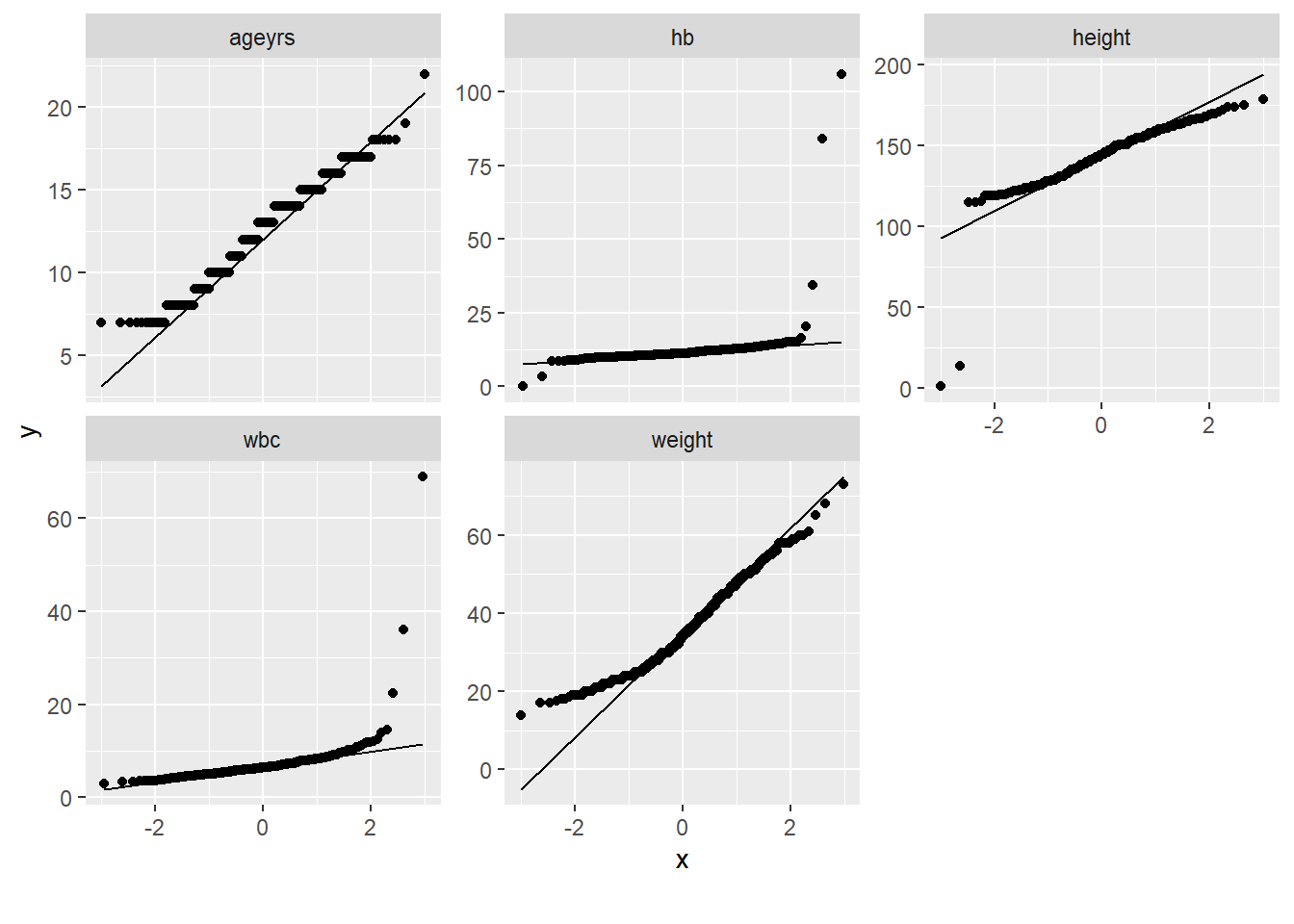
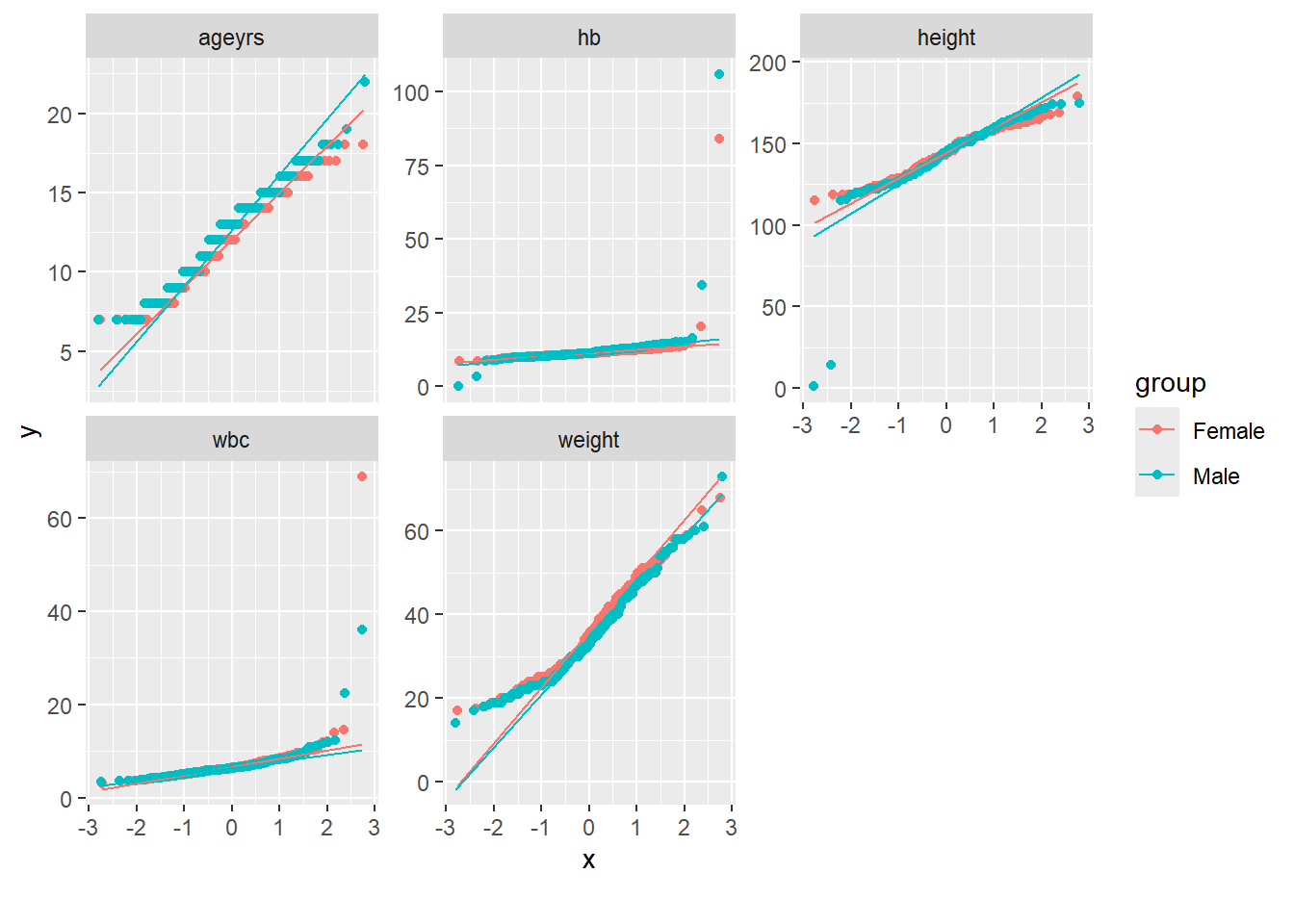
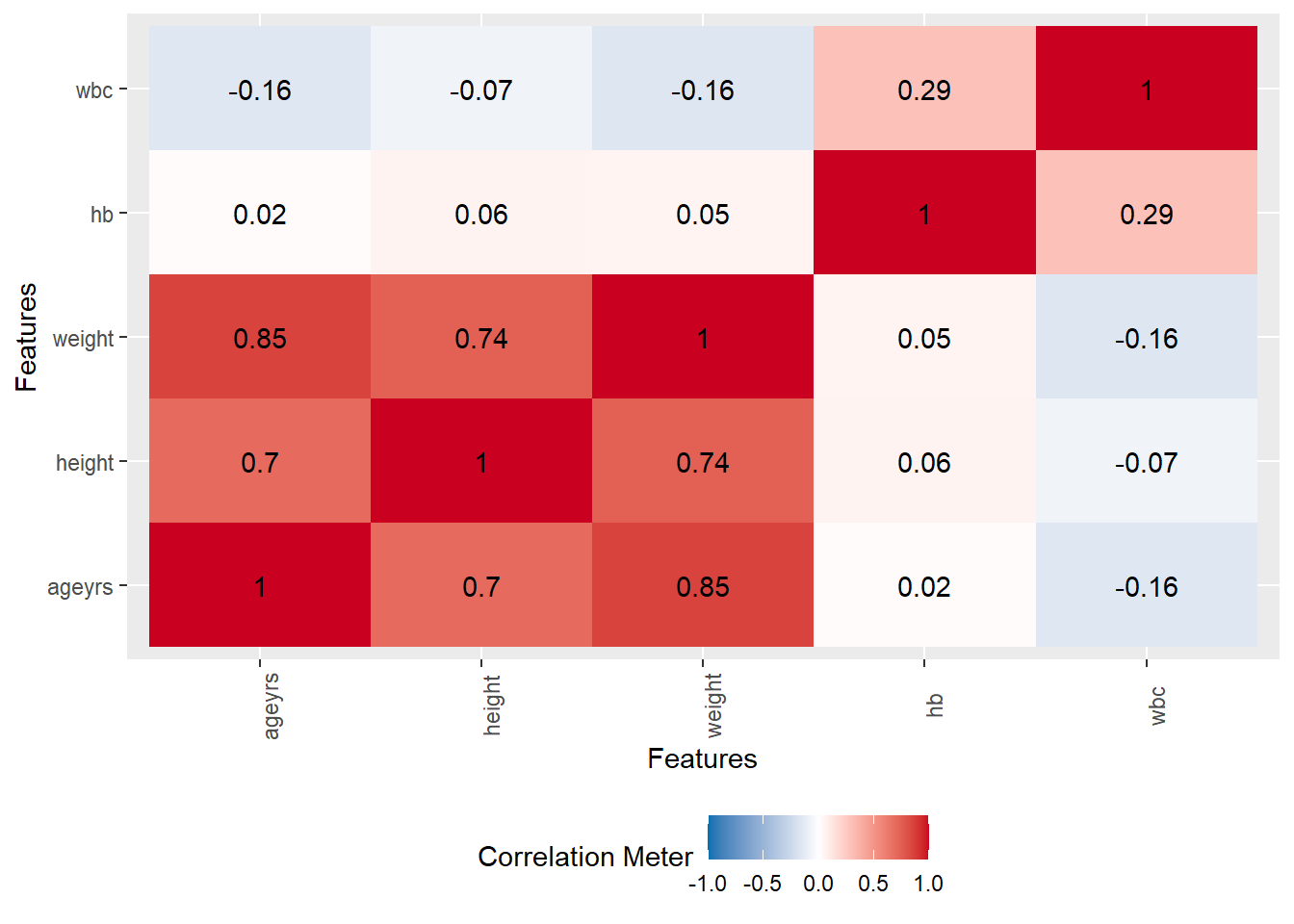
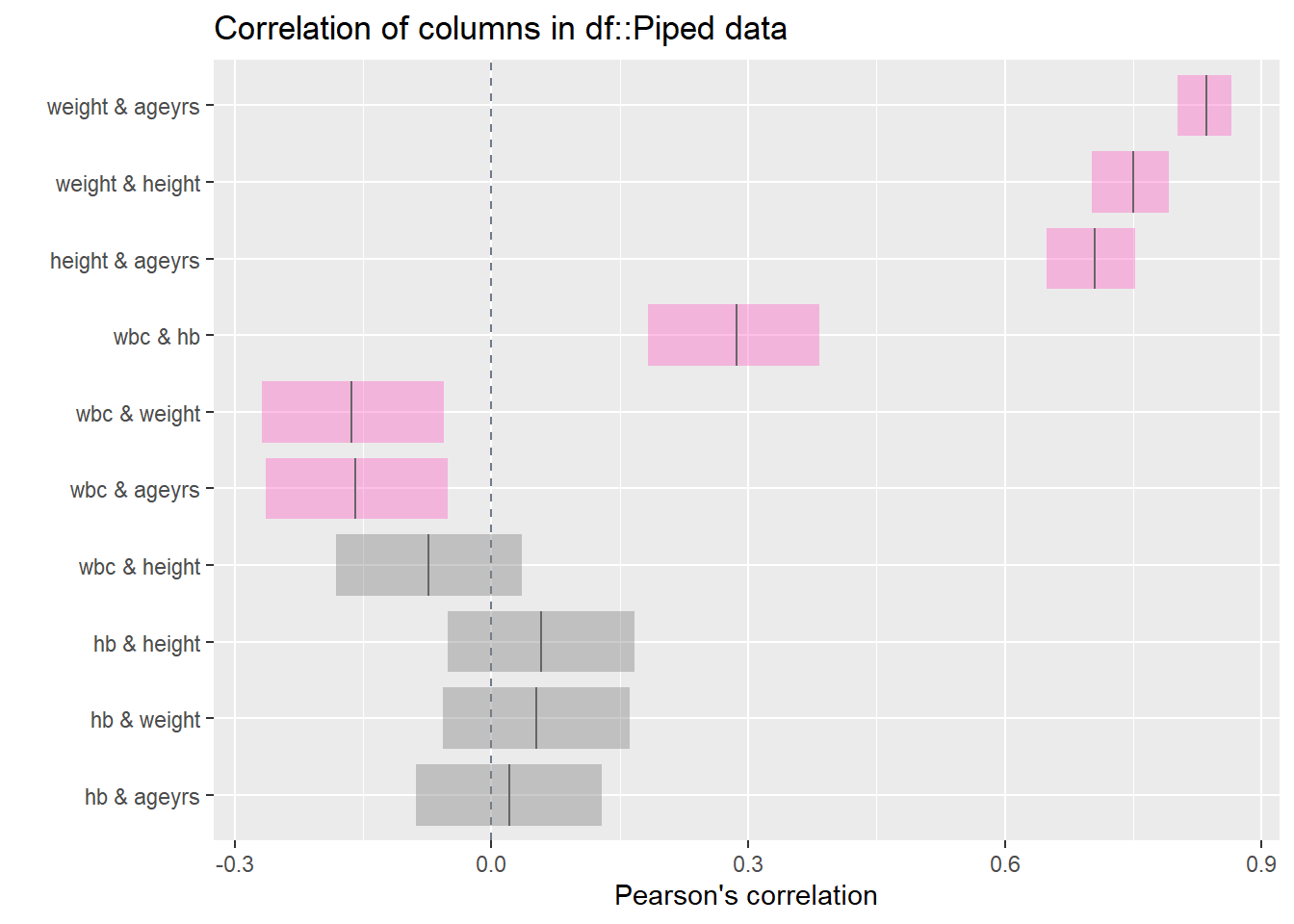
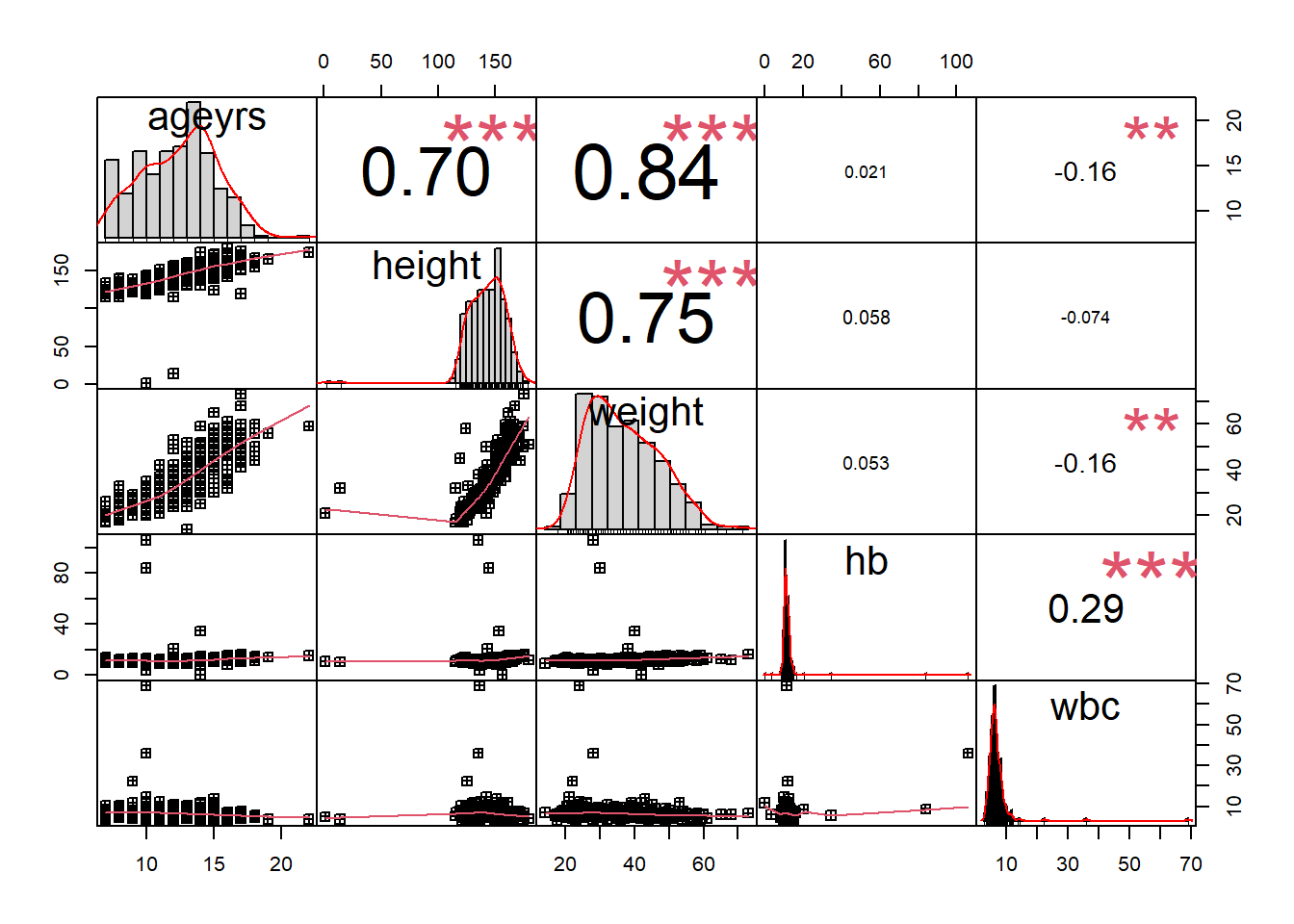
Rows: 367
Columns: 10
$ serial_no <fct> AB001, AB002, AB003, AB004, AB005, AB006, AB007, AB008, AB…
$ ageyrs <dbl> 7, 10, 18, 15, 12, 16, 10, 9, 14, 10, 8, 15, 16, 14, 10, 1…
$ sex <fct> Male, Male, Female, Male, Male, Male, Male, Female, Male, …
$ religion <fct> Christianity, Christianity, Christianity, Christianity, Ch…
$ educ_status <fct> Primary, Primary, JSS, JSS, JSS, Primary, Primary, Primary…
$ height <dbl> 124, 128, 160, 155, 115, 154, 135, 130, 140, 120, 129, 161…
$ weight <dbl> 22, 23, 44, 45, 32, 40, 28, 26, 30, 20, 25, 53, 56, 32, 27…
$ hb <dbl> 11.3, 12.5, 11.5, 12.8, NA, 11.1, 106.0, 11.1, 11.5, 11.4,…
$ wbc <dbl> 4.3, 5.3, 5.6, 5.5, NA, 6.6, 36.0, 11.8, 8.9, 7.6, 6.6, 12…
$ schisto <fct> nil, +++, nil, nil, nil, nil, +++, nil, ++, nil, +, +++, n…