Code
rm(list = ls(all = TRUE))
dat <-
foreign::read.dta("C:/Dataset/bea_organ_damage_28122013.dta")
dataF <-
readstata13::read.dta13(
"C:/Dataset/olivia_data_wide.dta",
nonint.factors = TRUE)First we read in the data
rm(list = ls(all = TRUE))
dat <-
foreign::read.dta("C:/Dataset/bea_organ_damage_28122013.dta")
dataF <-
readstata13::read.dta13(
"C:/Dataset/olivia_data_wide.dta",
nonint.factors = TRUE)Next we select three variables for plotting, keep only the complete cases and then store the ggplot() into an object called BP.
BP <-
dat %>%
select(q12weight, q2idtype, q3sex) %>%
na.omit() %>%
ggplot(aes(x = q2idtype, y = q12weight, fill = q3sex))Next we draw our boxplot with axis labels, title, axes format, and color specification.
BP +
geom_boxplot() +
theme_test() +
labs(title="My Boxplot", x="Case or Control", y="Weight (hgs)") +
theme(plot.title = element_text(size=15, face="bold"),
axis.text.x = element_text(size=12),
axis.text.y = element_text(size=12),
axis.title.x = element_text(size=13),
axis.title.y = element_text(size=13)) +
scale_color_discrete(name = "Sex")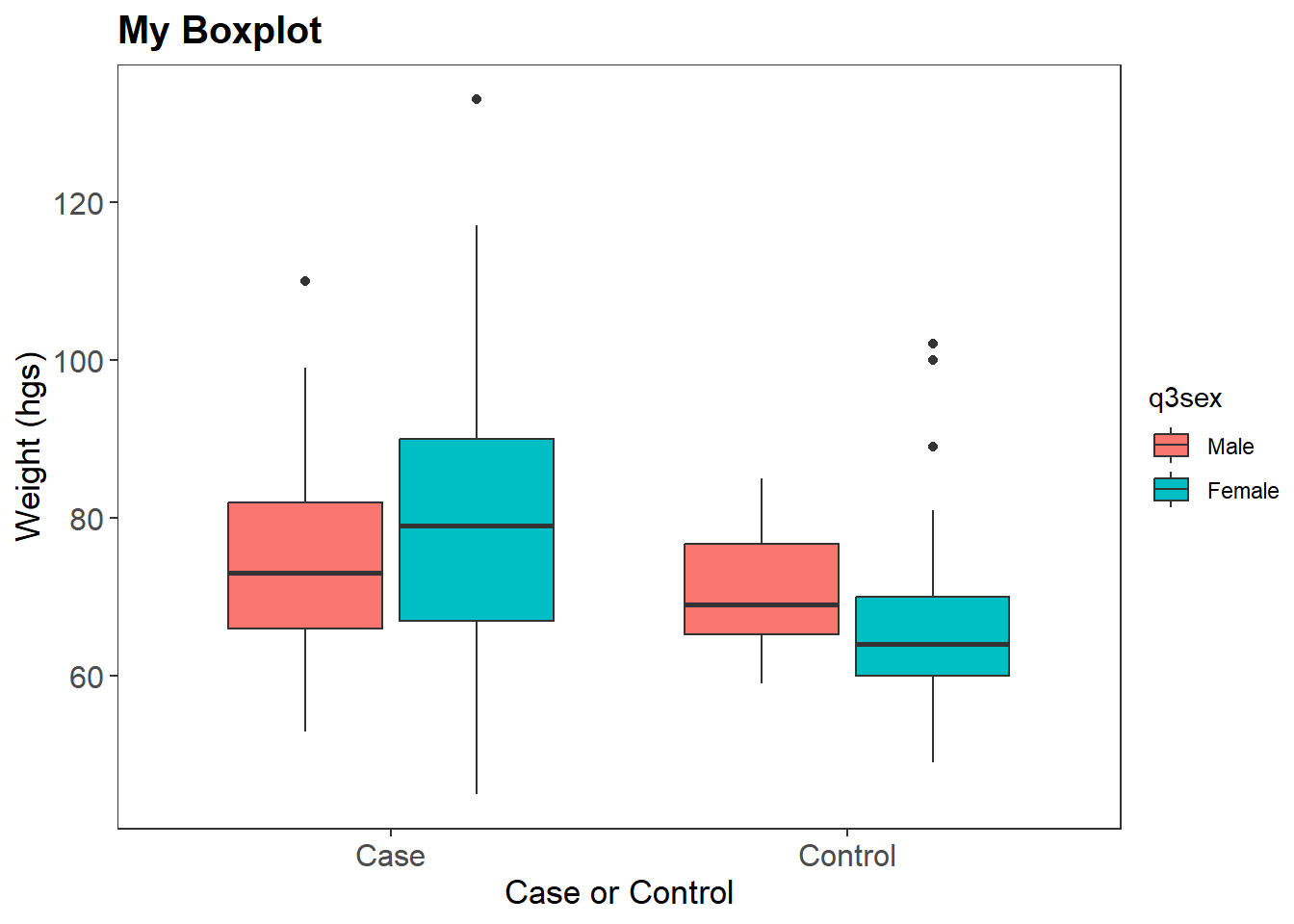
Newt we set up a similar boxplot but this time use the color option for the ggplot() and not the fill option.
BP <-
dat %>%
select(q12weight, q2idtype, q3sex) %>%
na.omit() %>%
ggplot(aes(x = q2idtype, y = q12weight, color = q3sex))BP +
geom_boxplot() +
theme_light() +
labs(title = "My Boxplot",
x = "Case or Control",
y = "Weight (hgs)") +
theme(plot.title=element_text(size=15, face="bold"),
axis.text.x=element_text(size=12),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=13),
axis.title.y=element_text(size=13)) +
scale_color_discrete(name="Sex")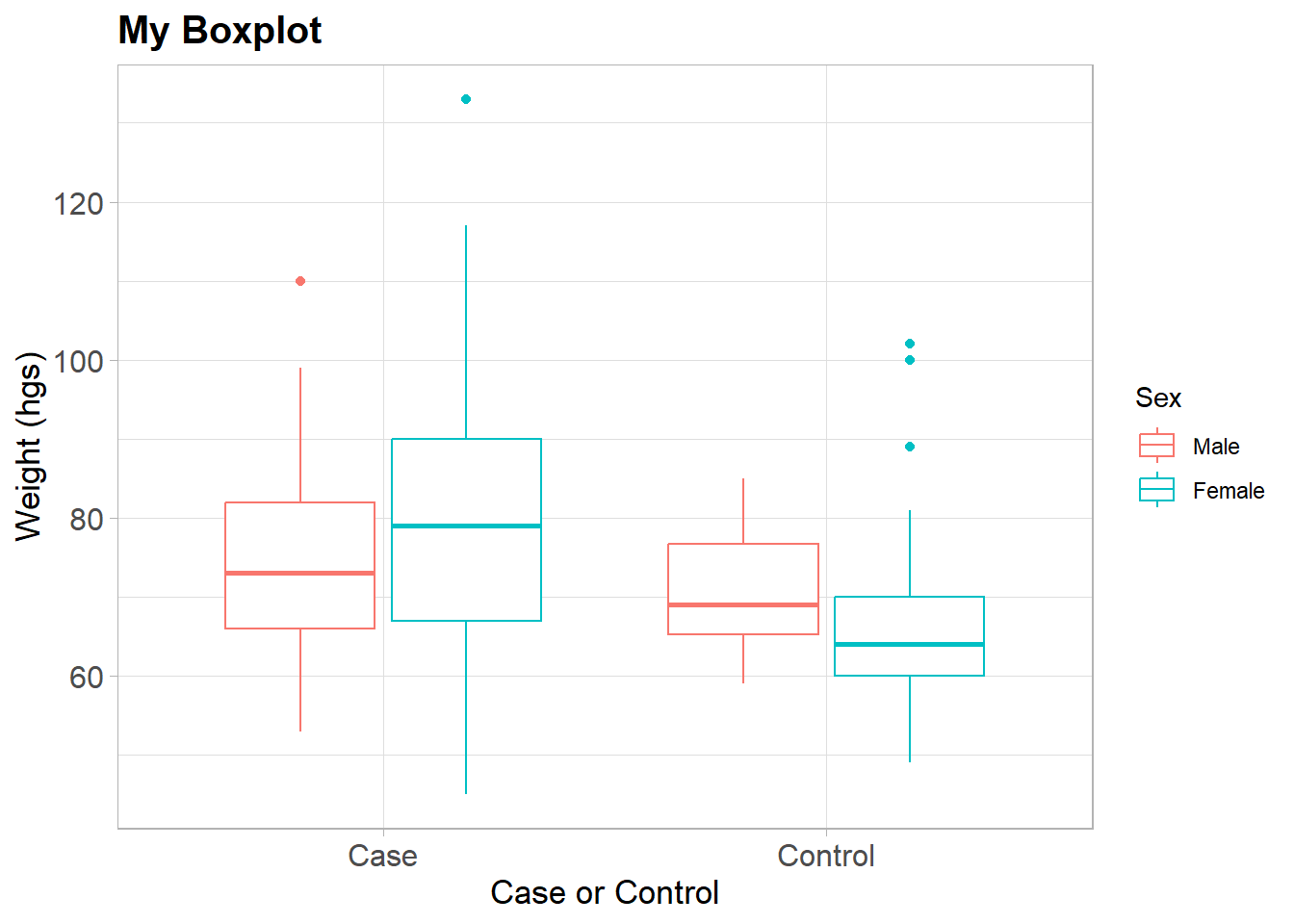
rm(BP, dat)Here we use a different dataset to draw the next boxplot
df1 <- read.csv("C:/Dataset/booking1.csv")Next we plot two boxplots on one graph
df1 %>%
na.omit() %>%
ggplot(aes(x = sex)) +
geom_boxplot(aes(y = weight, color="red")) +
geom_boxplot(aes(y = height, color = "steelblue")) +
labs(color = "Anthropometrics") +
scale_color_manual(labels = c("Weight","Height"),
values = c("red", "steelblue"))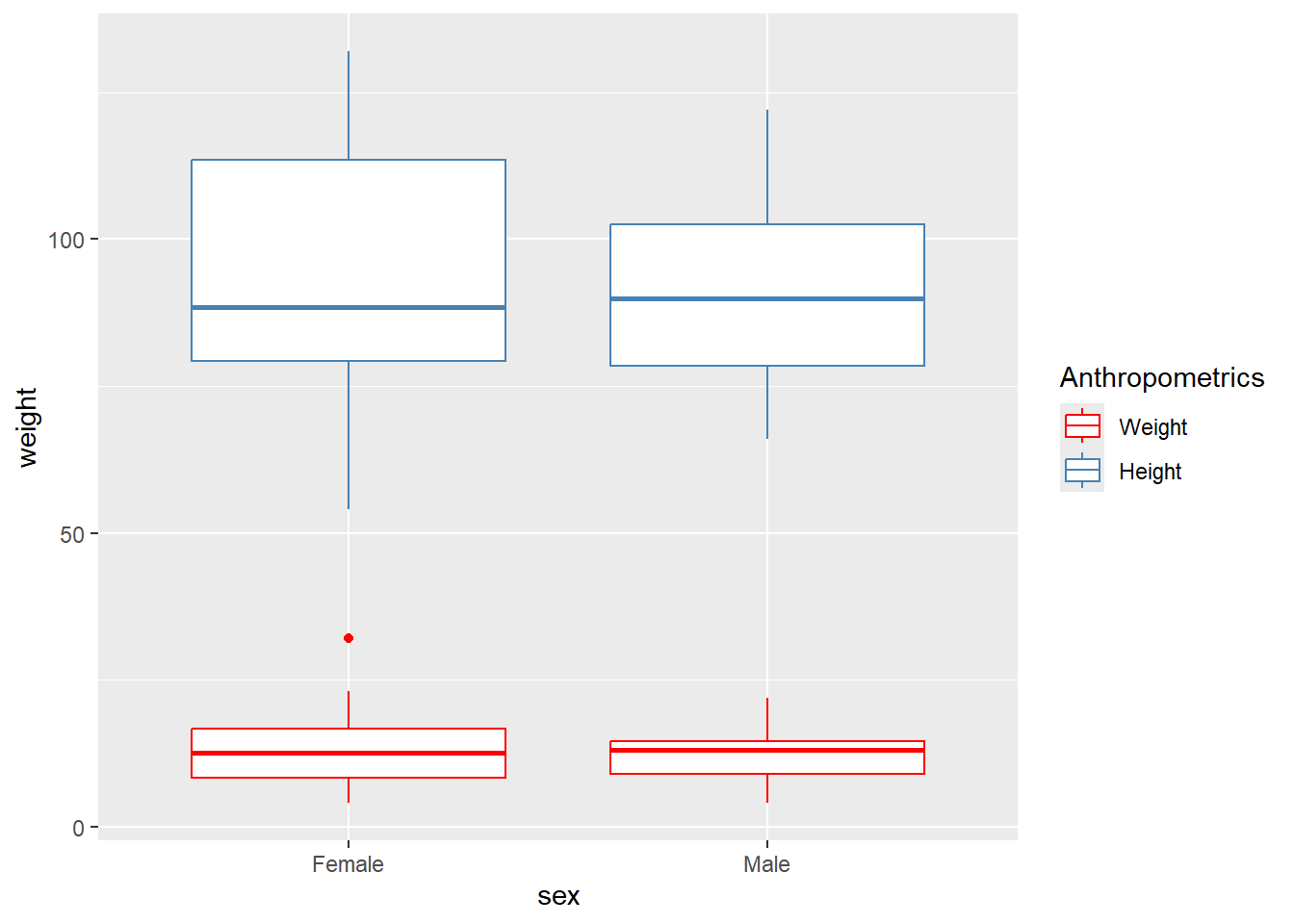
rm(df1)We then use the ToothGrowth data for for the next few boxplots
data(ToothGrowth)
ToothGrowth <-
ToothGrowth %>%
mutate(dose = factor(dose))
p <-
ToothGrowth %>%
ggplot(aes(x = dose, y = len))And then we form the ggplot object
Other renditions of the boxplot is as shown below. First rotated one
p + geom_boxplot() + coord_flip() # Axis rotated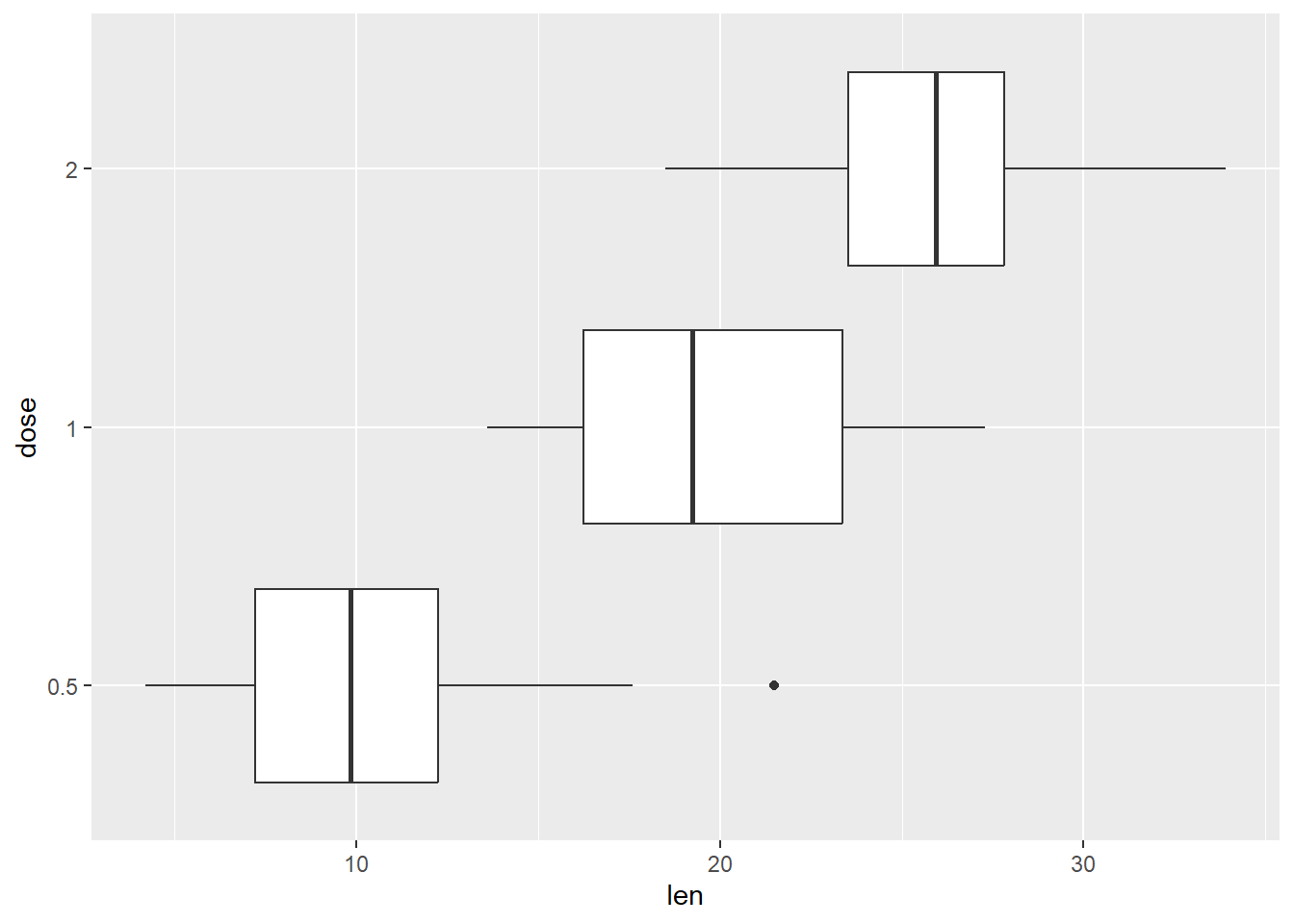
Notched boxplot
p + geom_boxplot(notch=TRUE)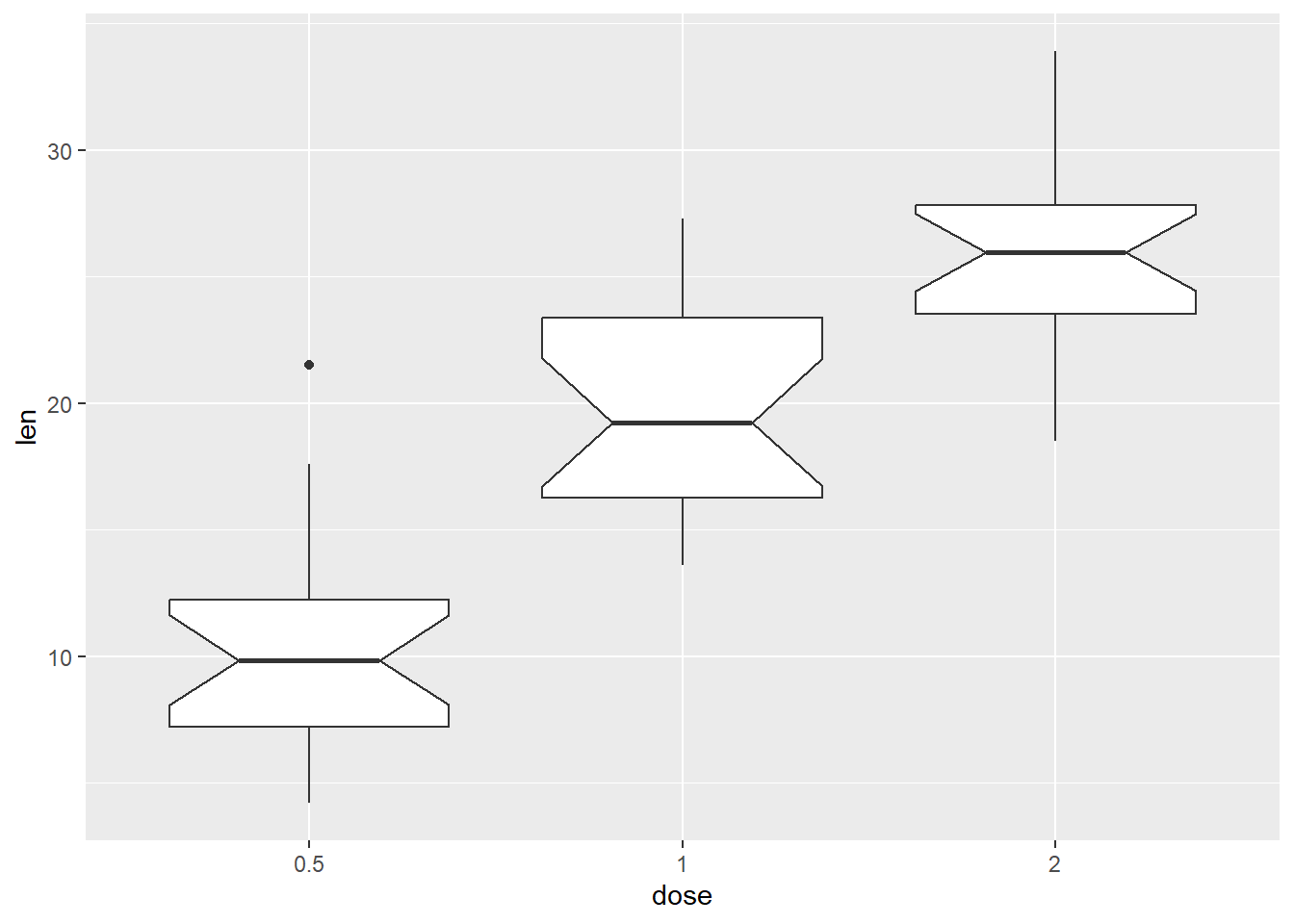
Customizaton of the outlier
p + geom_boxplot(
outlier.colour="red", outlier.shape=8, outlier.size=4)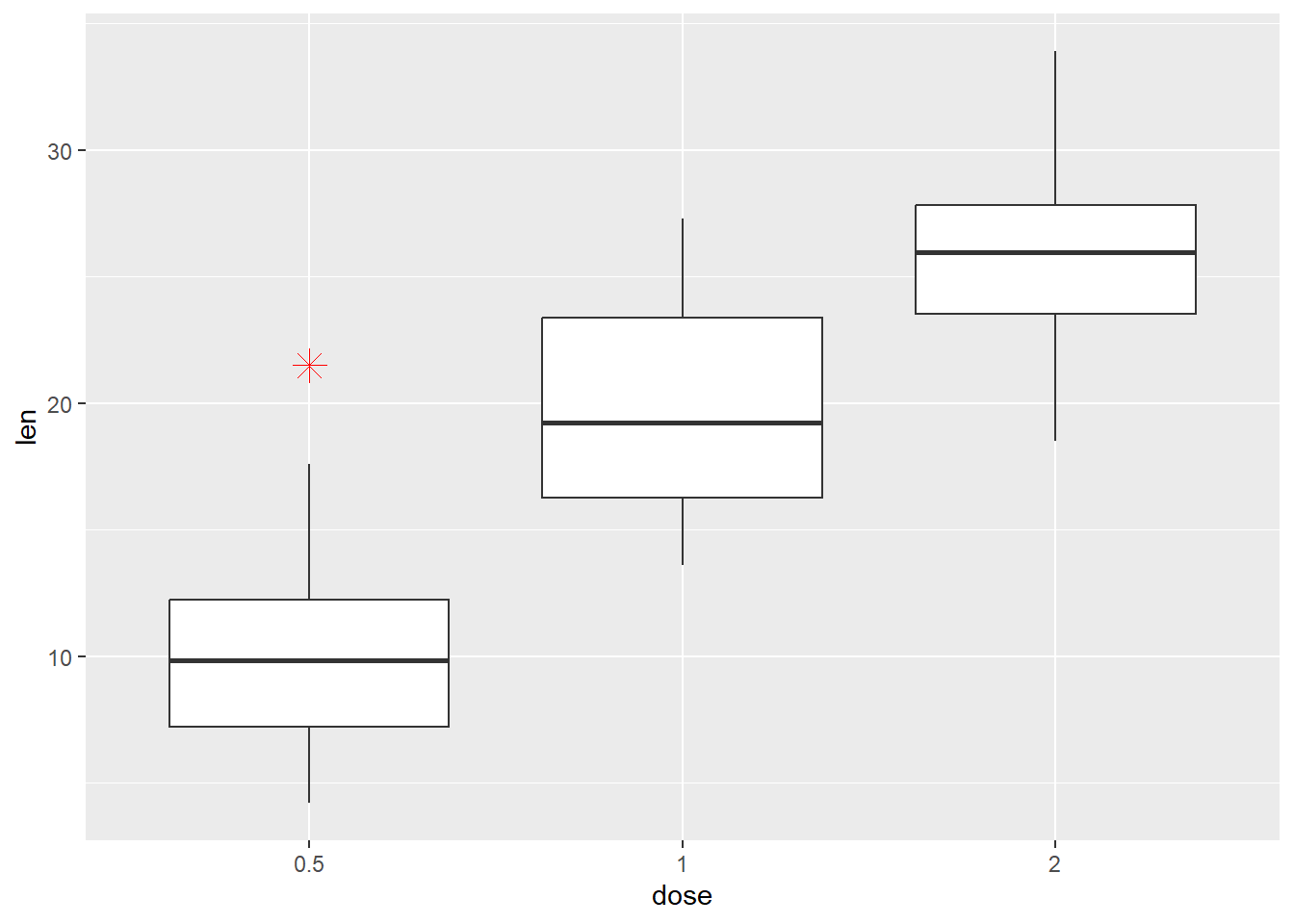
We add a statistic to the plot here
p + geom_boxplot() +
stat_summary(
fun = mean, geom = "point", shape = 18, size = 4, col = "red")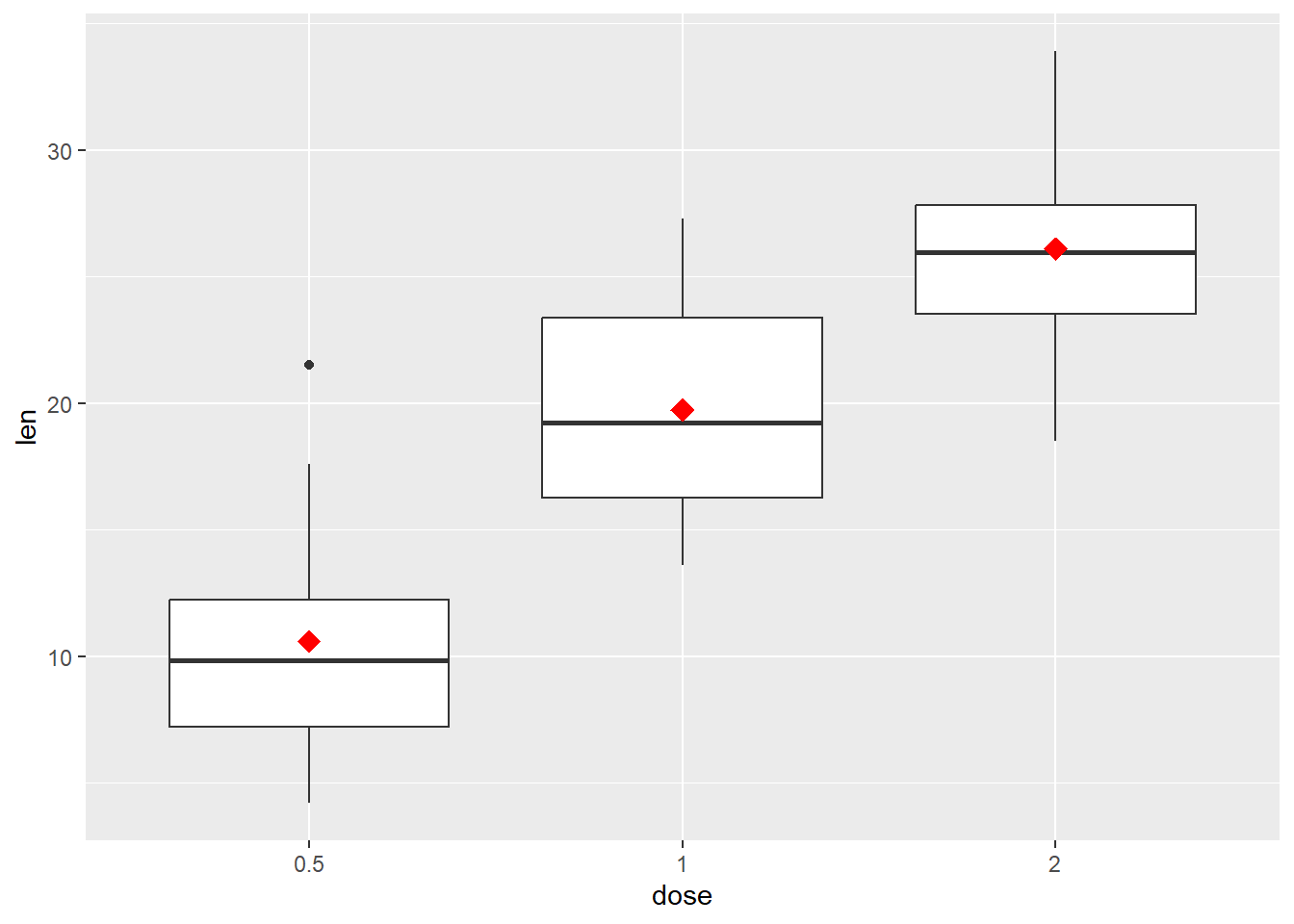
And then limit the categories the x axis
p +
geom_boxplot() +
scale_x_discrete(limits=c("0.5", "2"))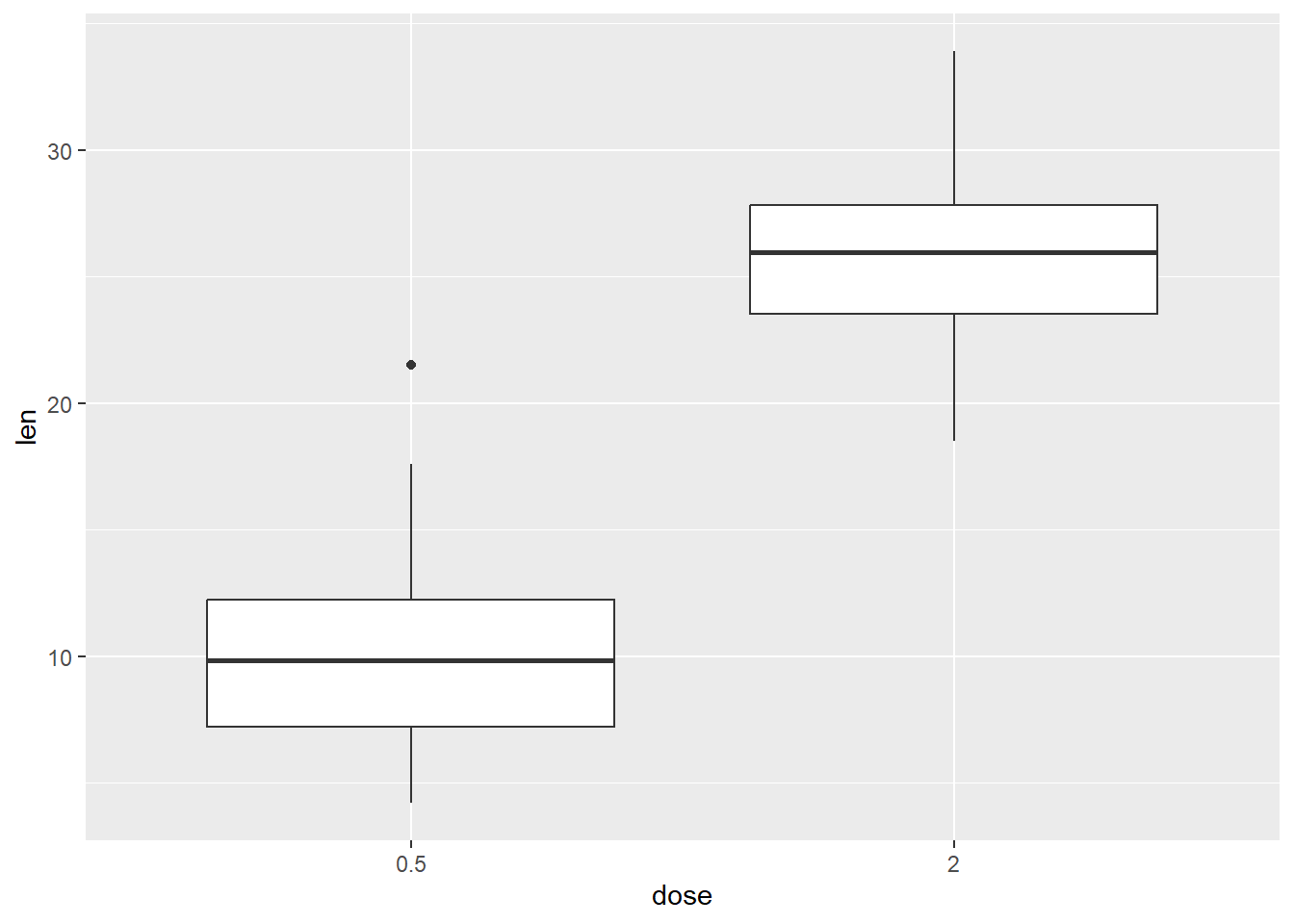
Next a boxplot with a superimposed dotplot
p +
geom_boxplot() +
geom_dotplot(
binaxis='y', stackdir='center', dotsize=0.5, binwidth = 1, col = "red") 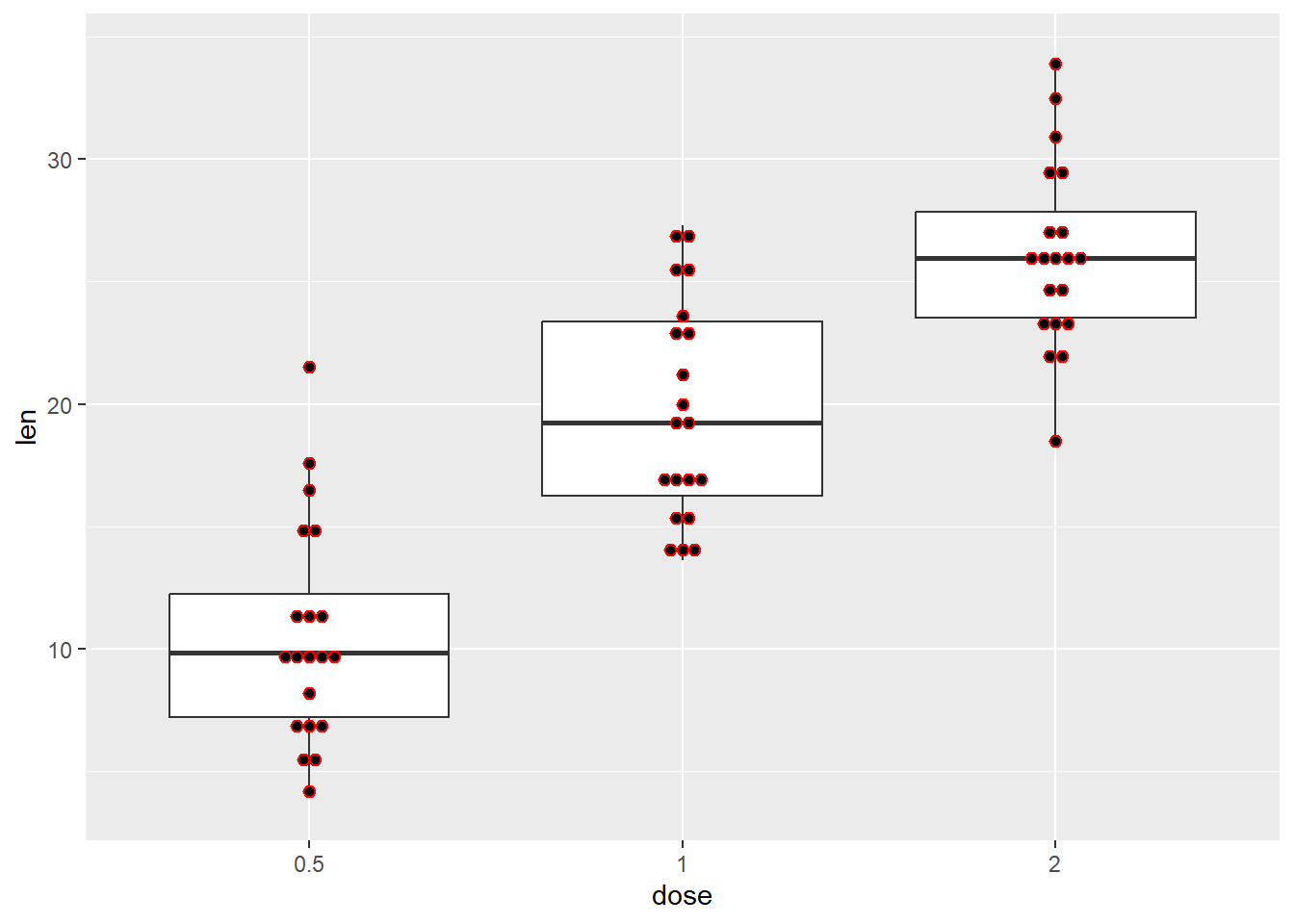
And a boxplot with superimposed jittered points
p +
geom_boxplot() +
geom_jitter(shape=16, position=position_jitter(0.2))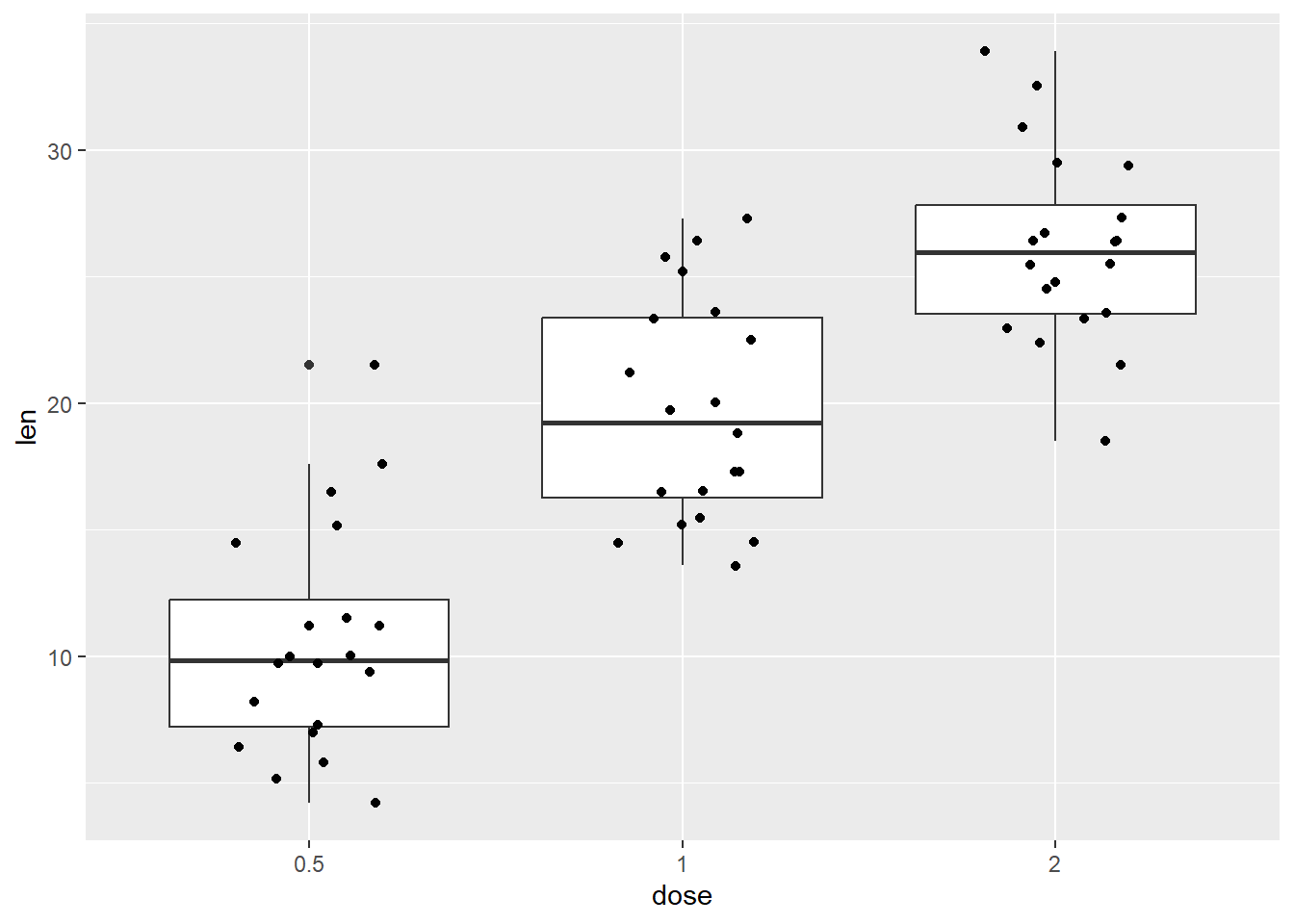
Next we manually set out own color scale
P <-
ToothGrowth %>%
ggplot(aes(factor(dose), len, color=dose))
P +
geom_boxplot() +
scale_color_manual(values=c("#999999", "#E69F00", "#56B4E9"))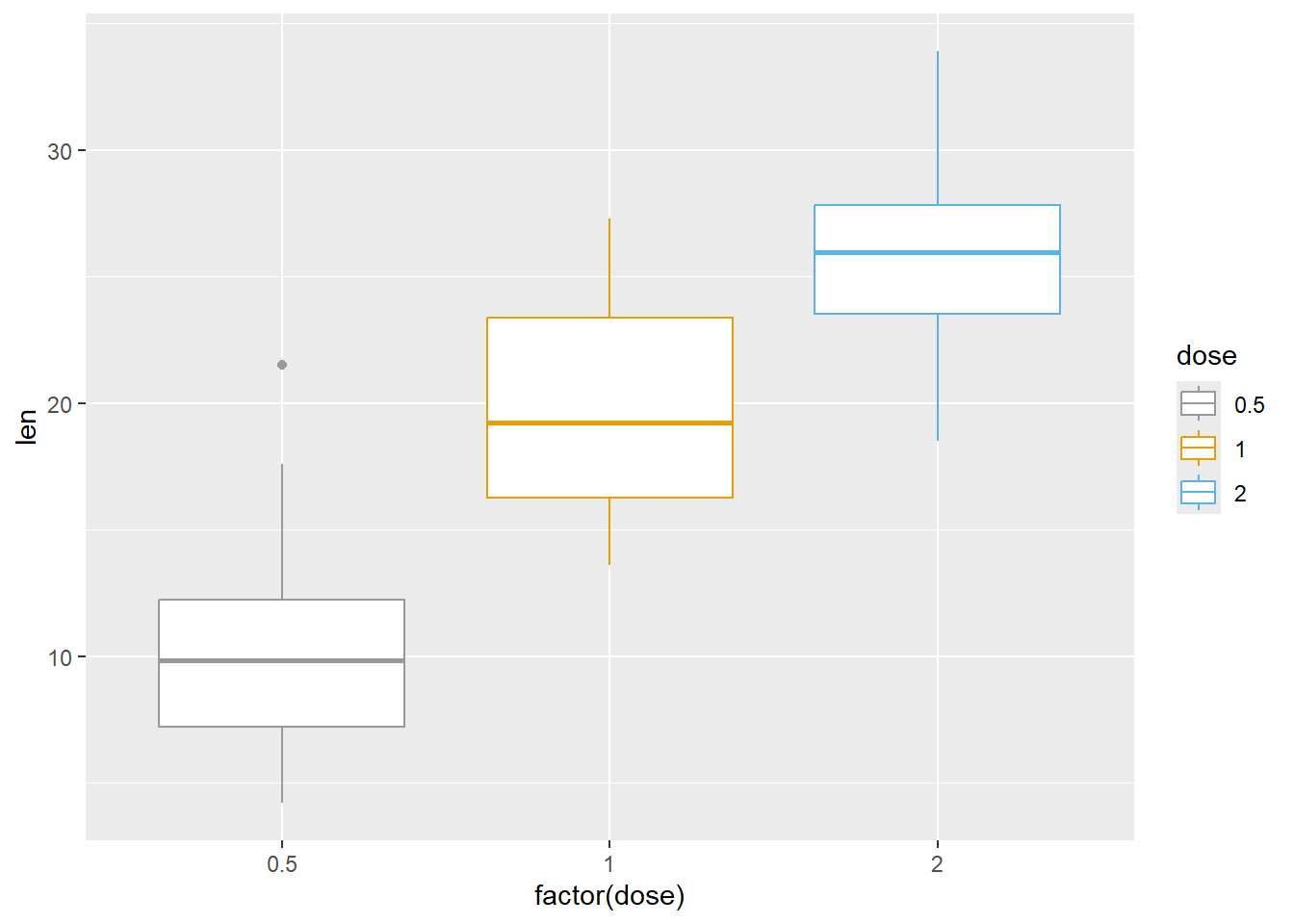
And use one of the color scales
P +
geom_boxplot() +
scale_color_brewer(palette="Dark2") +
scale_fill_brewer(palette="Dark2")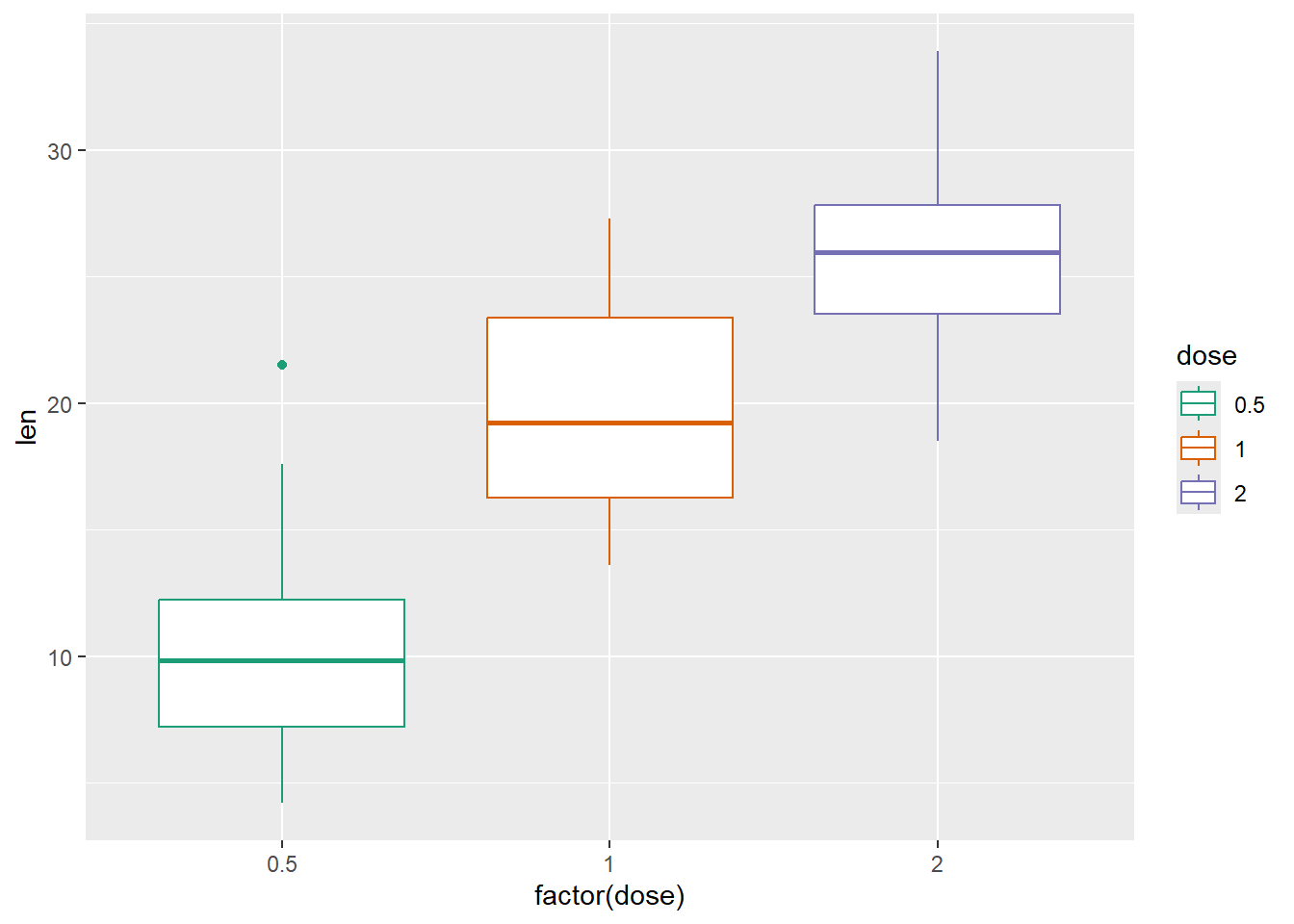
Change the legend position
P +
geom_boxplot() +
theme(legend.position = "top")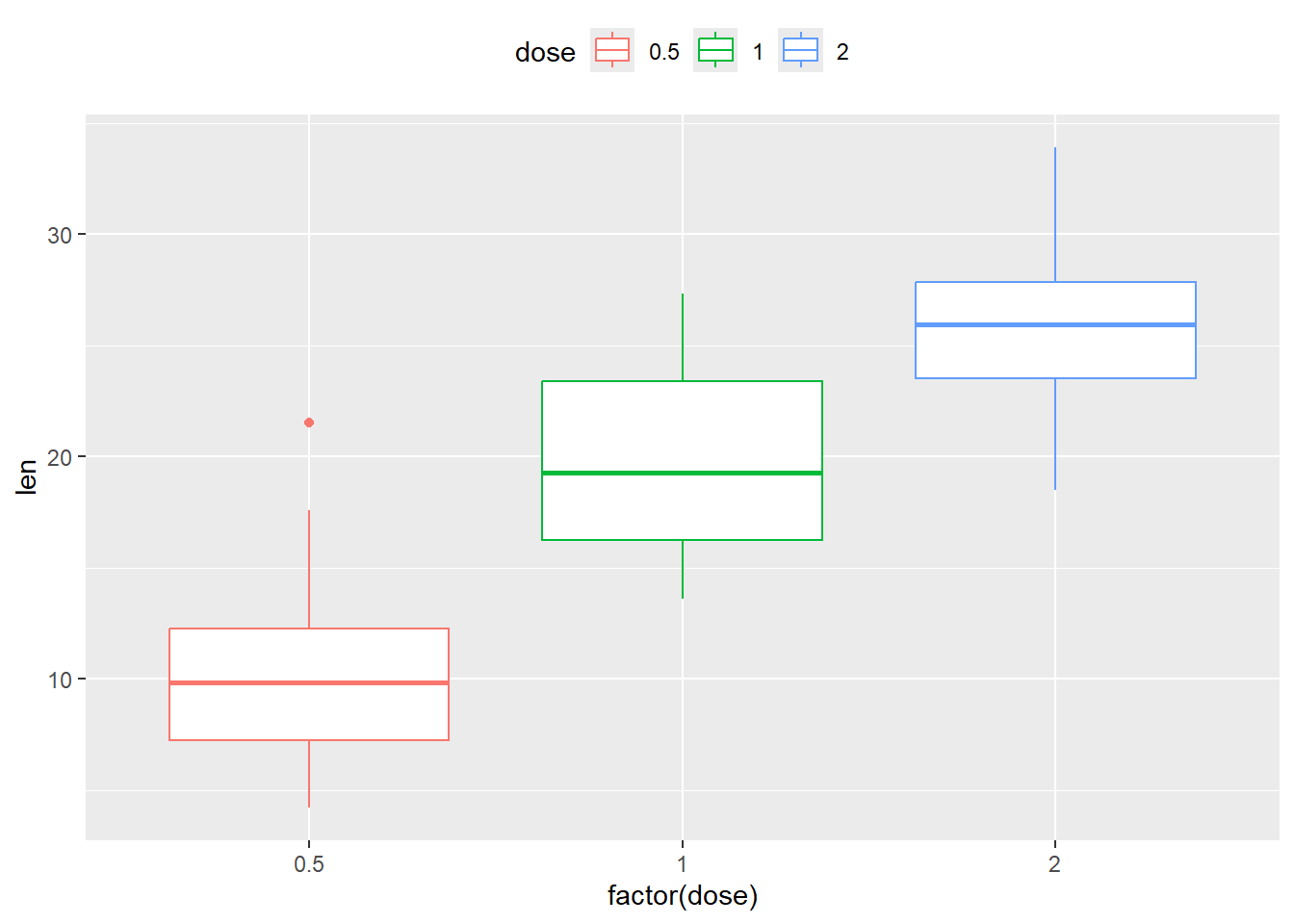
And remove legend
P +
geom_boxplot() +
theme(legend.position="none")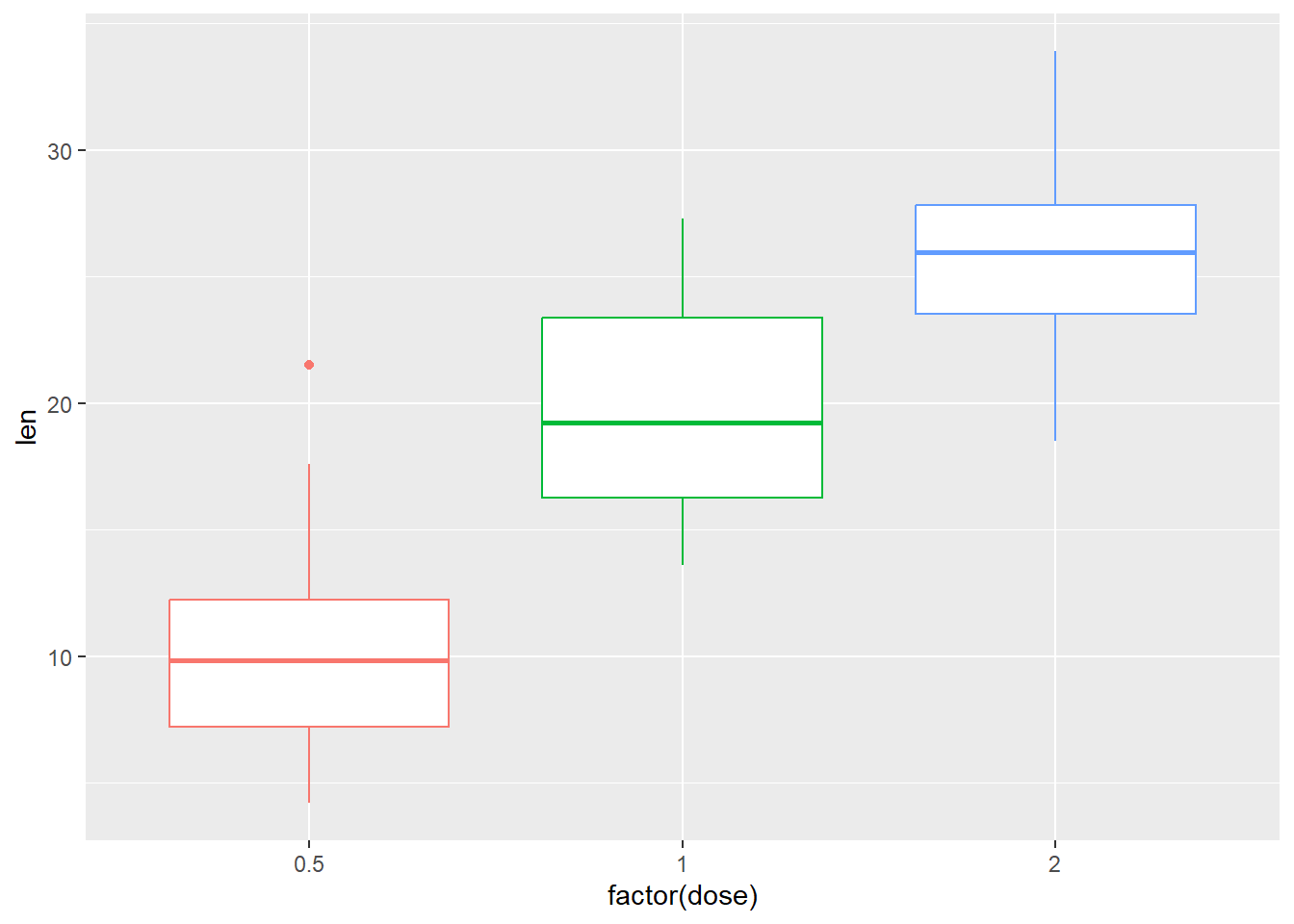
Change the order of items in the legend
P +
geom_boxplot() +
scale_x_discrete(limits=c("2", "0.5", "1"))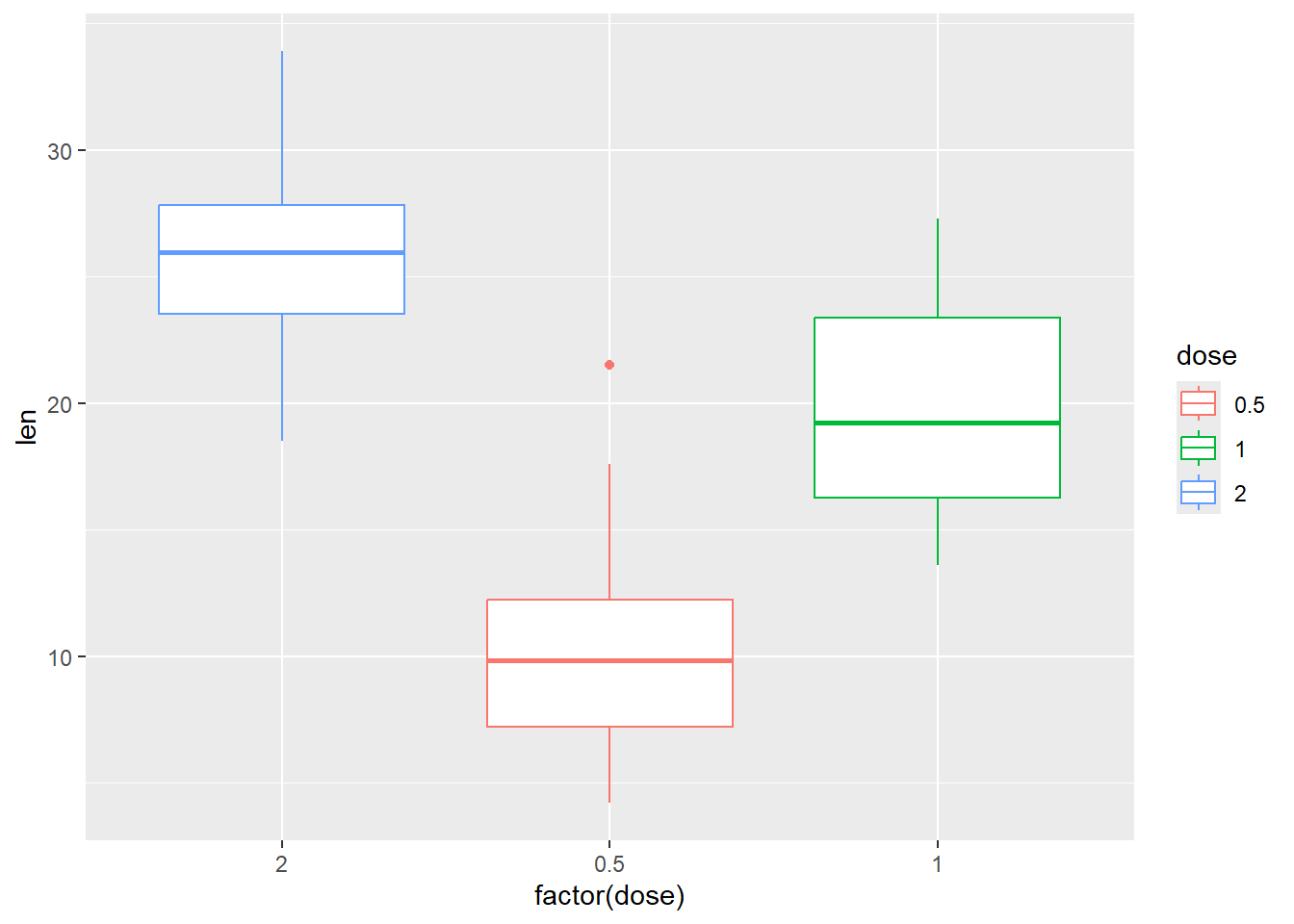
Change the position of the boxes
P <-
ggplot(ToothGrowth, aes(x=dose, y=len, fill=supp)) +
geom_boxplot(position=position_dodge(1))
P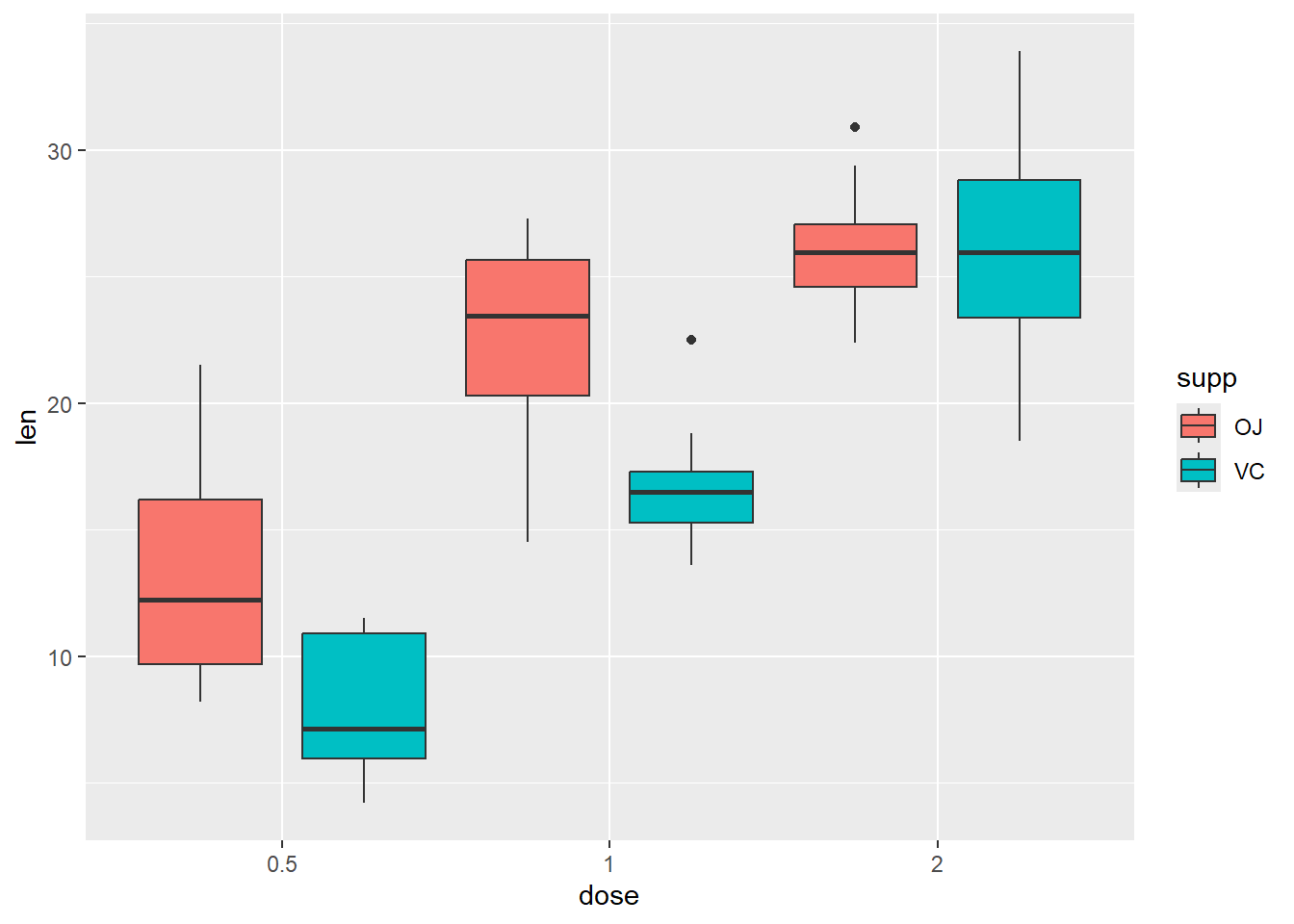
And then we add dots
P + geom_dotplot(binaxis='y', stackdir='center', position=position_dodge(1),
binwidth = 1)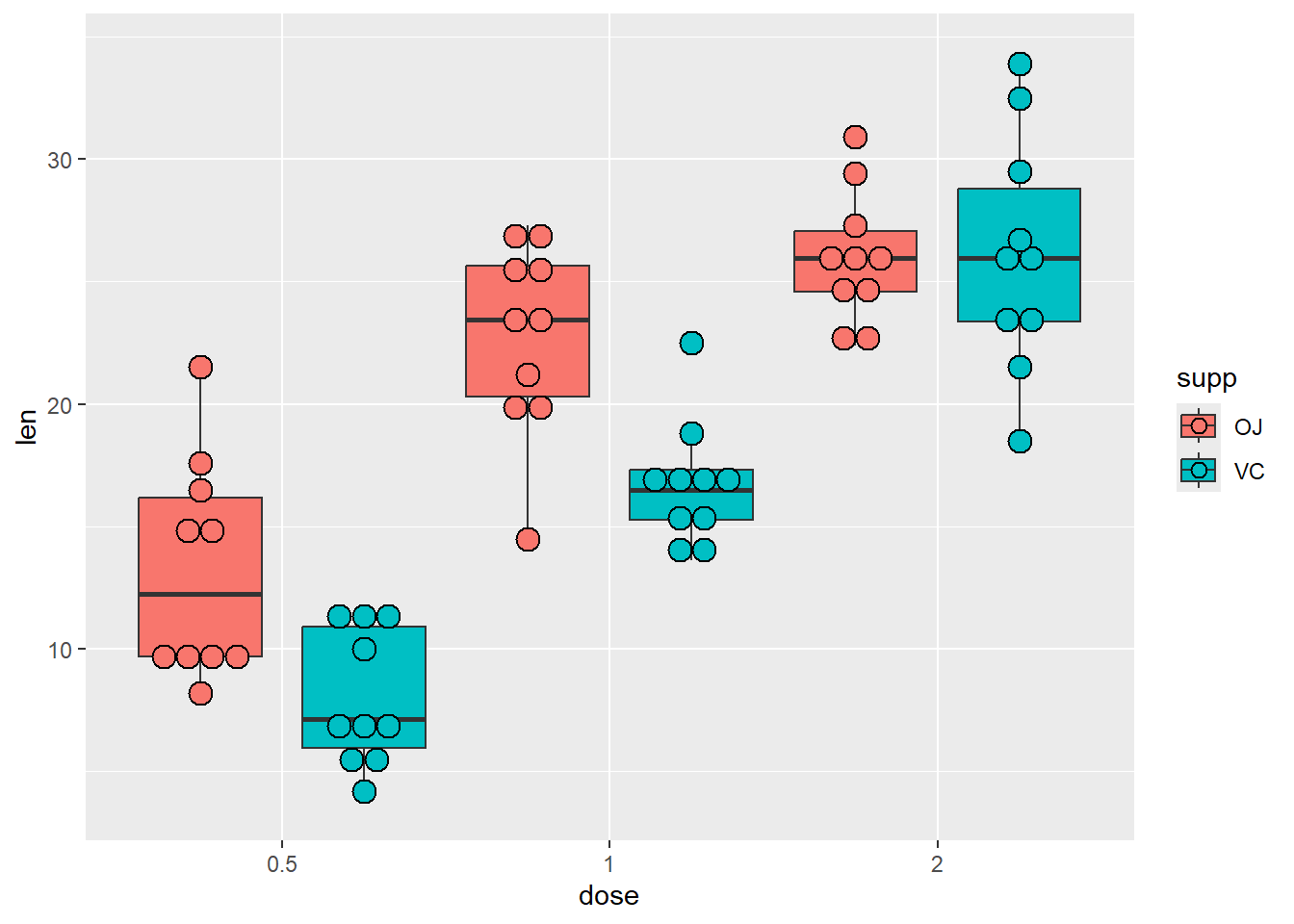
Customized
dataF %>%
select(mcv1, mcv2, mcv3, mcv4, mcv5, agecat, id) %>%
pivot_longer(cols = mcv1:mcv5, names_to = "Time", values_to = "MCV") %>%
ggplot(aes(x = Time, y = MCV, col = Time), fill = "snow1") +
geom_boxplot(
outlier.color = 'black',
outlier.shape = 23,
outlier.fill = "steelblue2",
outlier.size = 2) +
stat_summary(
aes(fill=Time),
fun.data = mean_se,
geom = "pointrange",
size=0.5,
shape =23,
color = "black",
show.legend = F) +
scale_color_manual(
name = "Measure",
values = c("red", "yellow", "green", "violet", "brown"),
labels = c("First","Second", "Third", "Fourth", "Fifth")) +
scale_x_discrete(
labels =c(
"mcv1" = "First MCV",
"mcv2" = "Second MCV",
"mcv3" = "Third MCV",
"mcv4" = "Fourth MCV",
"mcv5" = "Fifth MCV")) +
labs(title = "Distribution of MCVs over the review periods") +
theme(
plot.title = element_text(
family = "serif",
face = "bold.italic",
size = 14,
colour = "steelblue4",
hjust = 0.5))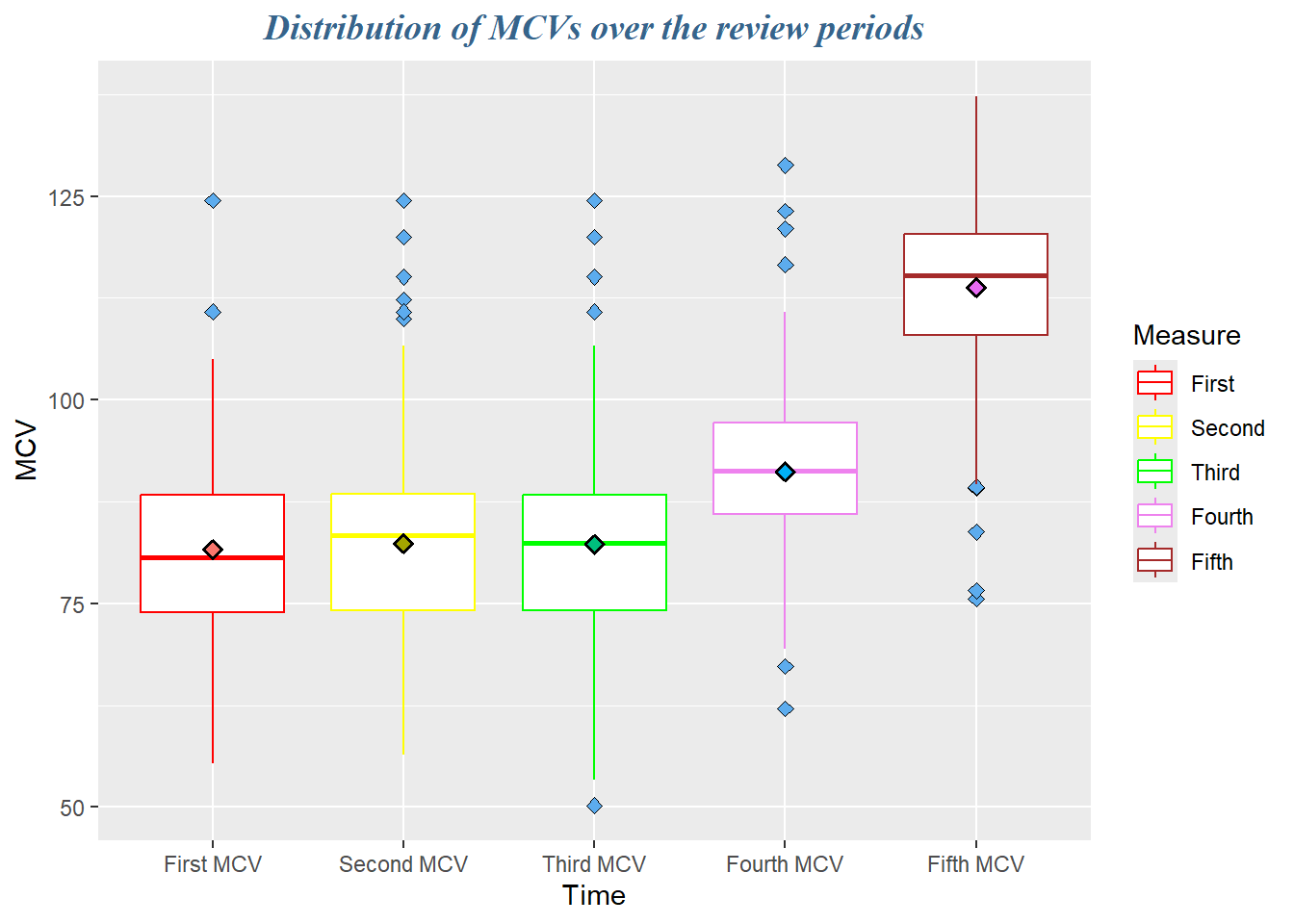
dataF %>%
select(mcv1, mcv2, mcv3, mcv4, mcv5, agecat, id) %>%
pivot_longer(cols = mcv1:mcv5, names_to = "Time", values_to = "MCV") %>%
ggplot(aes(x = Time, y = MCV, col = Time), fill = "snow1") +
geom_boxplot(outlier.color = "white", outlier.alpha = 0) +
geom_jitter(width =.2, alpha = .2, col=1) +
labs(
x = "Time of Sample taking",
y = "Mean Corpuscular Volume",
title = "Sequential changes in MCV over the study duration") +
theme_bw() +
scale_x_discrete(
labels = c(
"mcv1" = "First MCV",
"mcv2" = "Second MCV",
"mcv3" = "Third MCV",
"mcv4" = "Fourth MCV",
"mcv5" = "Fifth MCV"))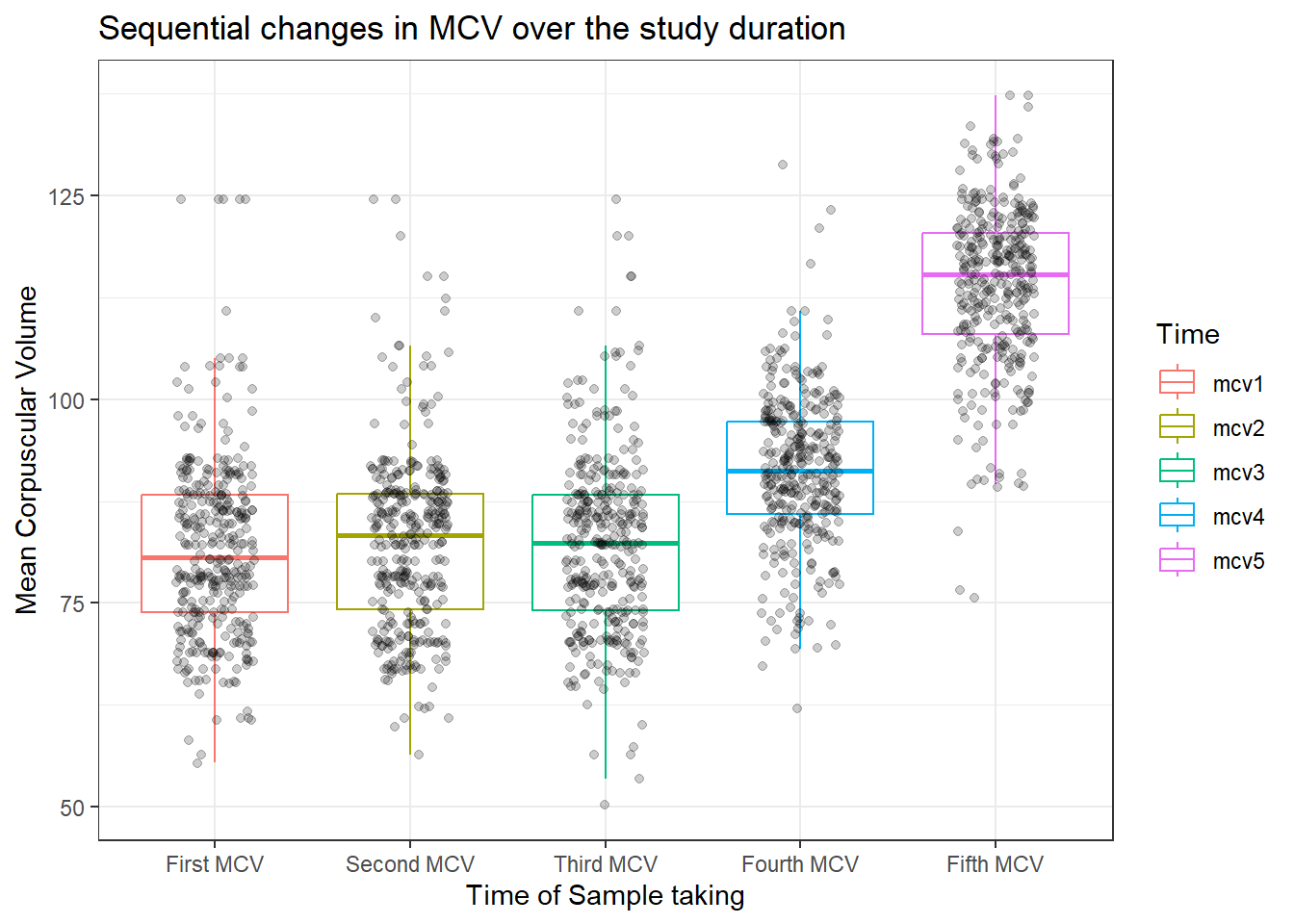
dataF %>%
select(mcv1, mcv2, mcv3, mcv4, mcv5, agecat, id) %>%
pivot_longer(cols = mcv1:mcv5, names_to = "Time", values_to = "MCV") %>%
ggplot(aes(x = Time, y = MCV, col = Time), fill = "snow1") +
ggbeeswarm::geom_beeswarm() +
labs(x = "Time of Sample taking",
y = "Mean Corpuscular Volume",
title = "Sequential changes in MCV over the study duration") +
theme_bw() +
scale_x_discrete(
labels = c(
"mcv1" = "First MCV", "mcv2" = "Second MCV", "mcv3" = "Third MCV",
"mcv4" = "Fourth MCV", "mcv5" = "Fifth MCV"))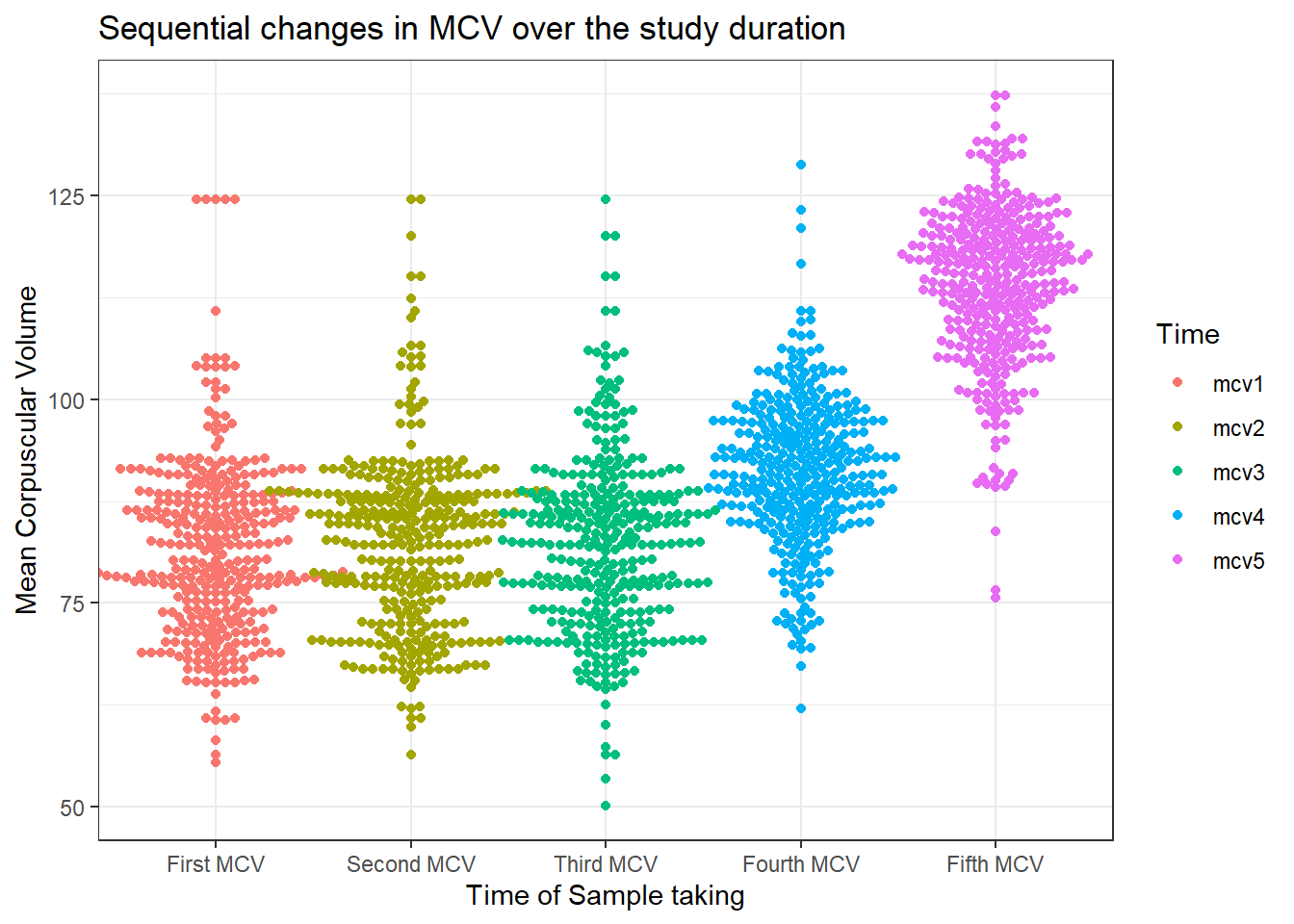
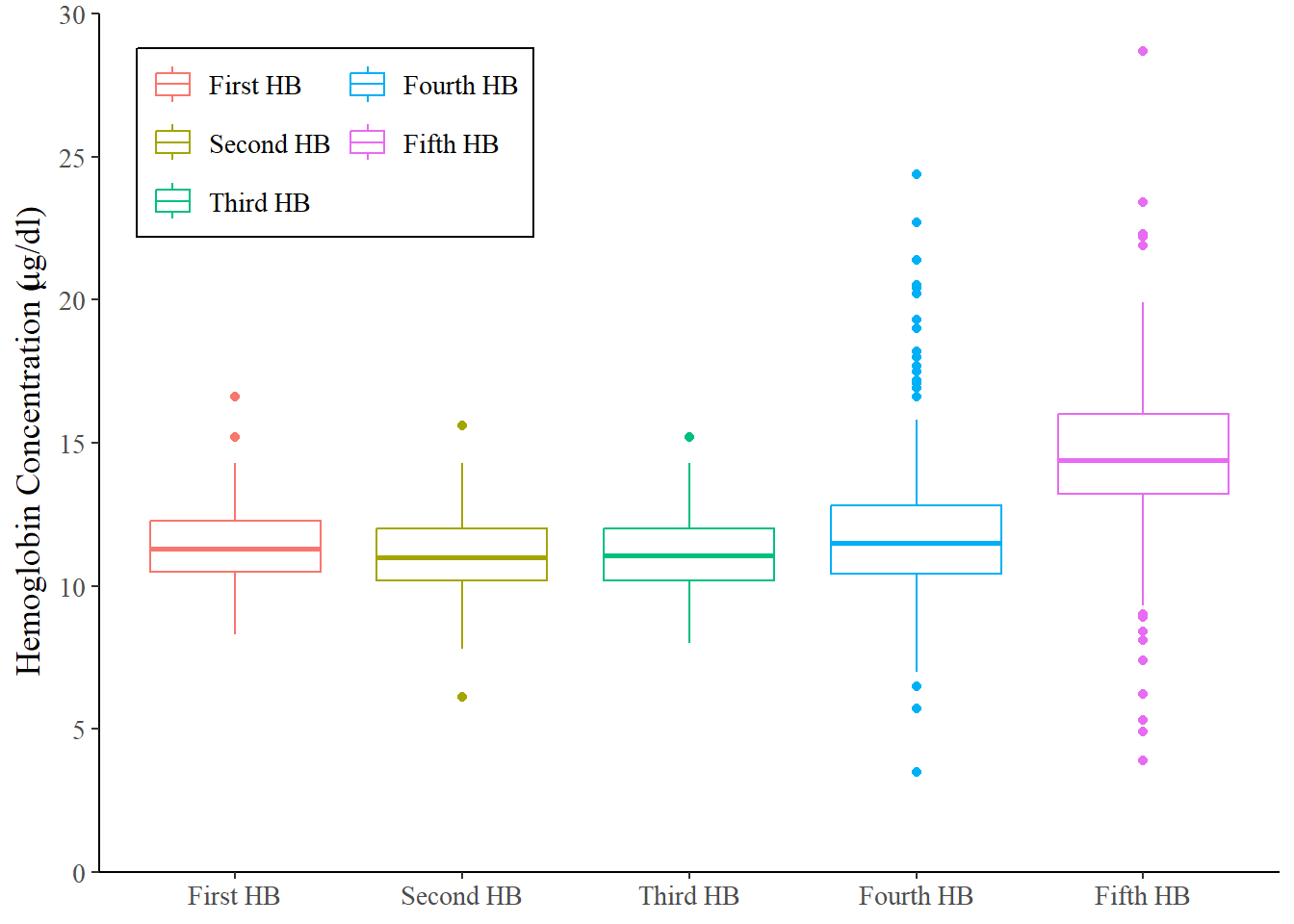
dataF %>%
select(hb1, hb2, hb3, hb4, hb5, agecat, id) %>%
pivot_longer(cols = hb1:hb5, names_to = "Time", values_to = "hb") %>%
ggplot(aes(x = Time, y = hb, color = Time)) +
geom_boxplot()+
scale_x_discrete(
name = NULL,
labels = c(
"hb1" = "First HB", "hb2" = "Second HB",
"hb3" = "Third HB", "hb4" = "Fourth HB",
"hb5" = "Fifth HB"))+
scale_y_continuous(
name = expression(paste('Hemoglobin Concentration (', mu, 'g/dl)')),
limits = c(0, 30),
breaks = seq(0, 30, 5),
expand = c(0,0))+
scale_color_discrete(
name = NULL,
labels = c(
"hb1" = "First HB", "hb2" = "Second HB",
"hb3" = "Third HB", "hb4" = "Fourth HB",
"hb5" = "Fifth HB"))+
guides(color=guide_legend(ncol=2,title = NULL))+
theme_classic()+
theme(
text = element_text(family = "serif", size = 13),
legend.background = element_rect(color = "black"),
legend.position = "inside",
legend.position.inside = c(0.2, 0.85),
legend.direction = "horizontal")