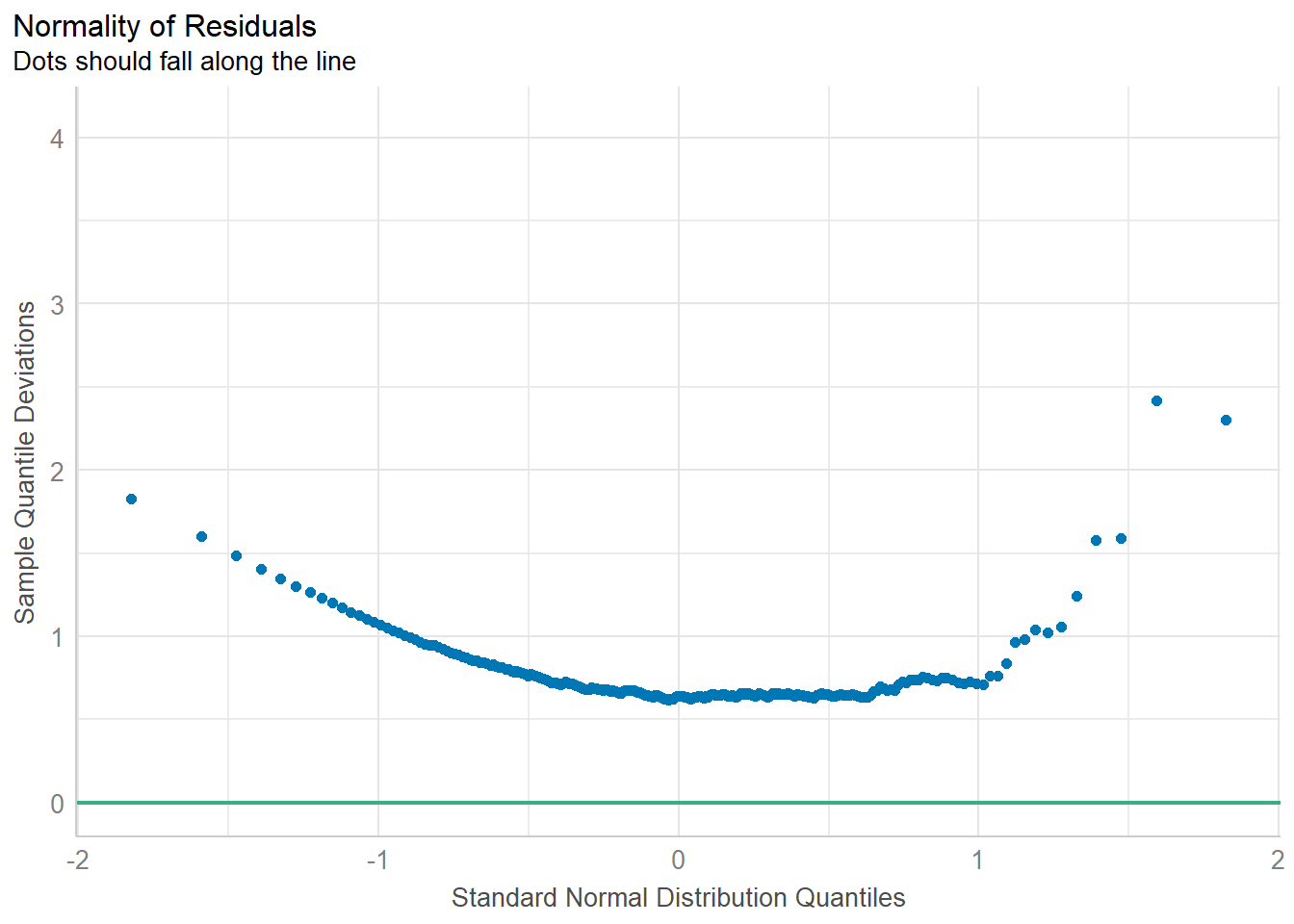
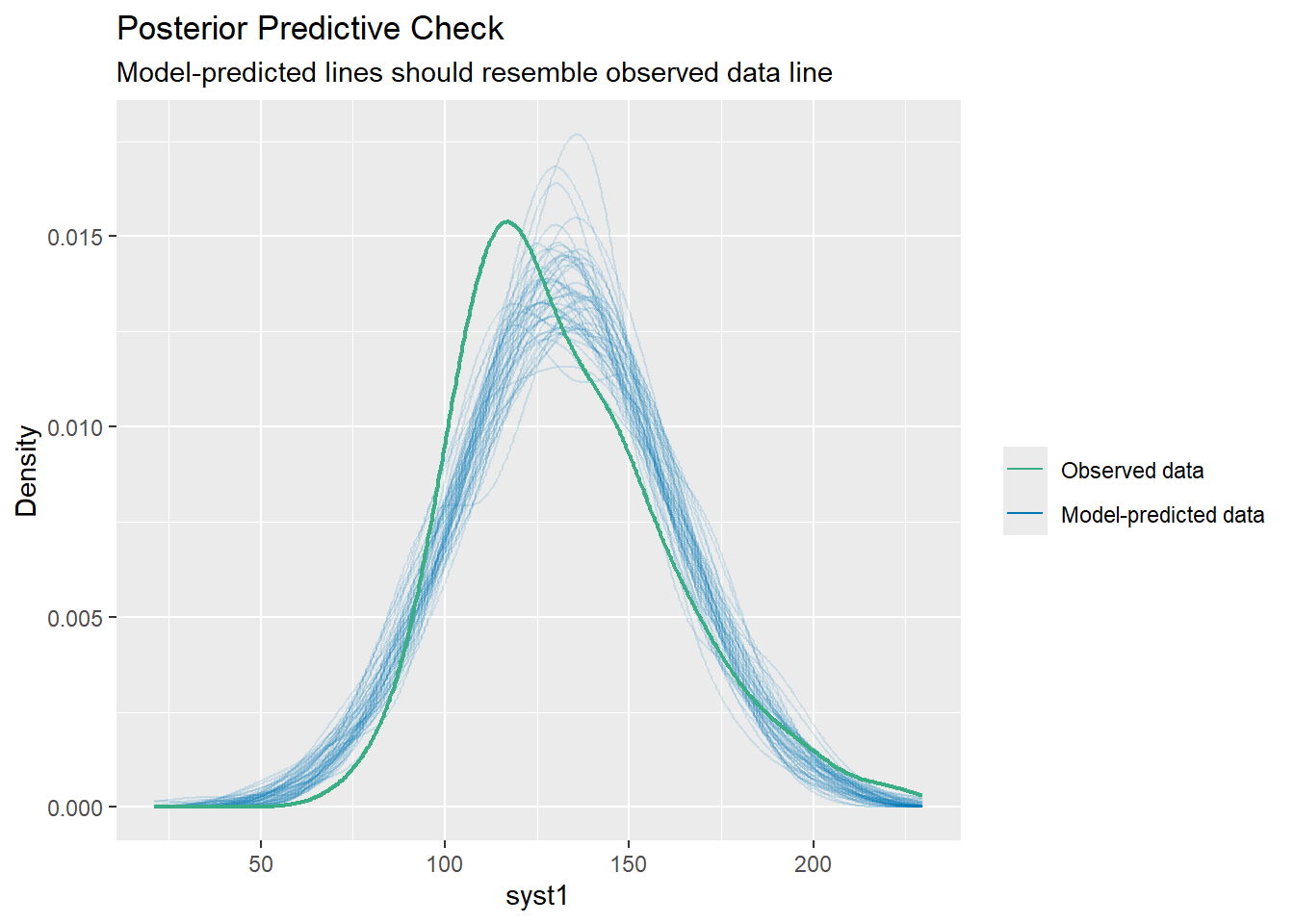
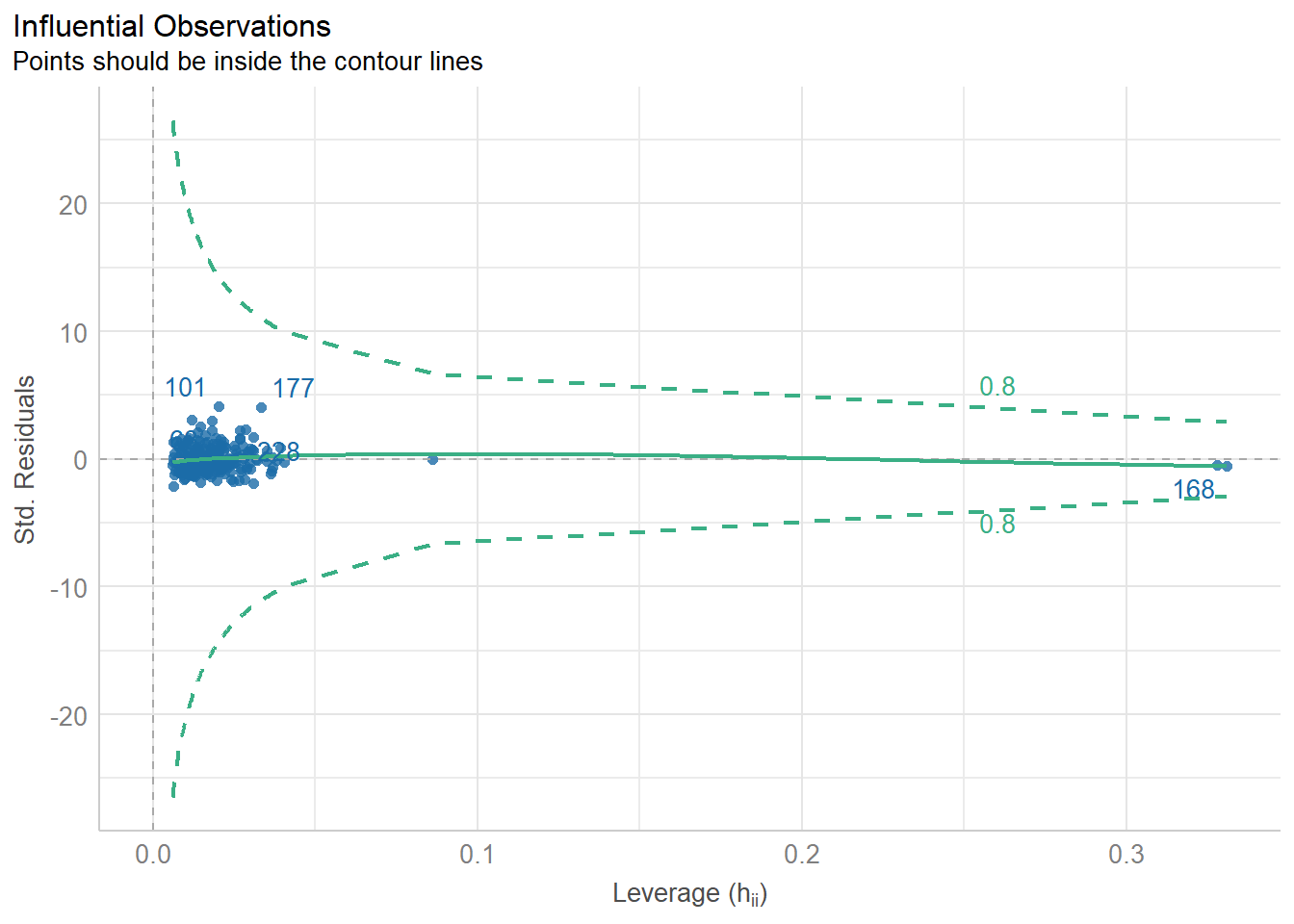
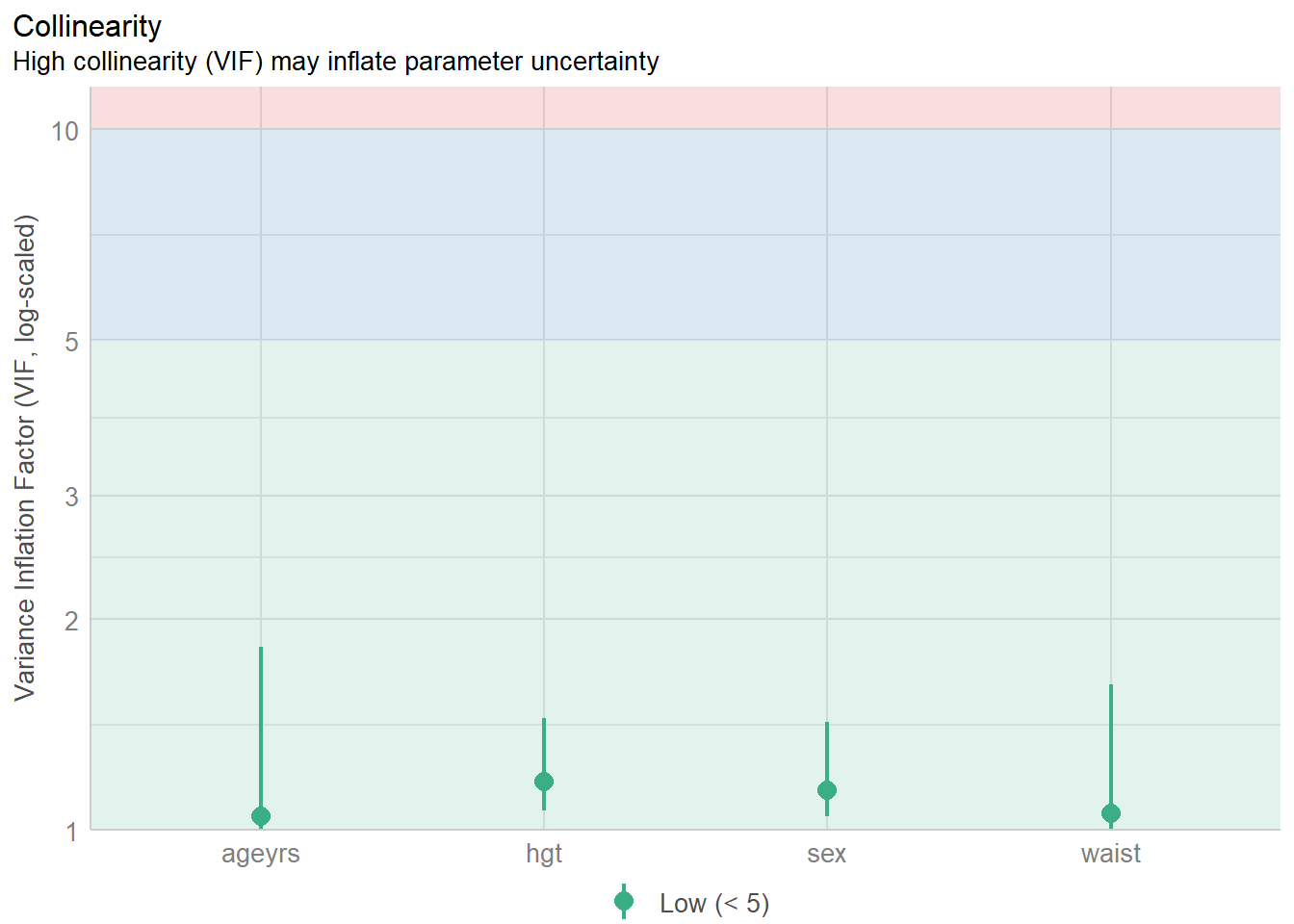
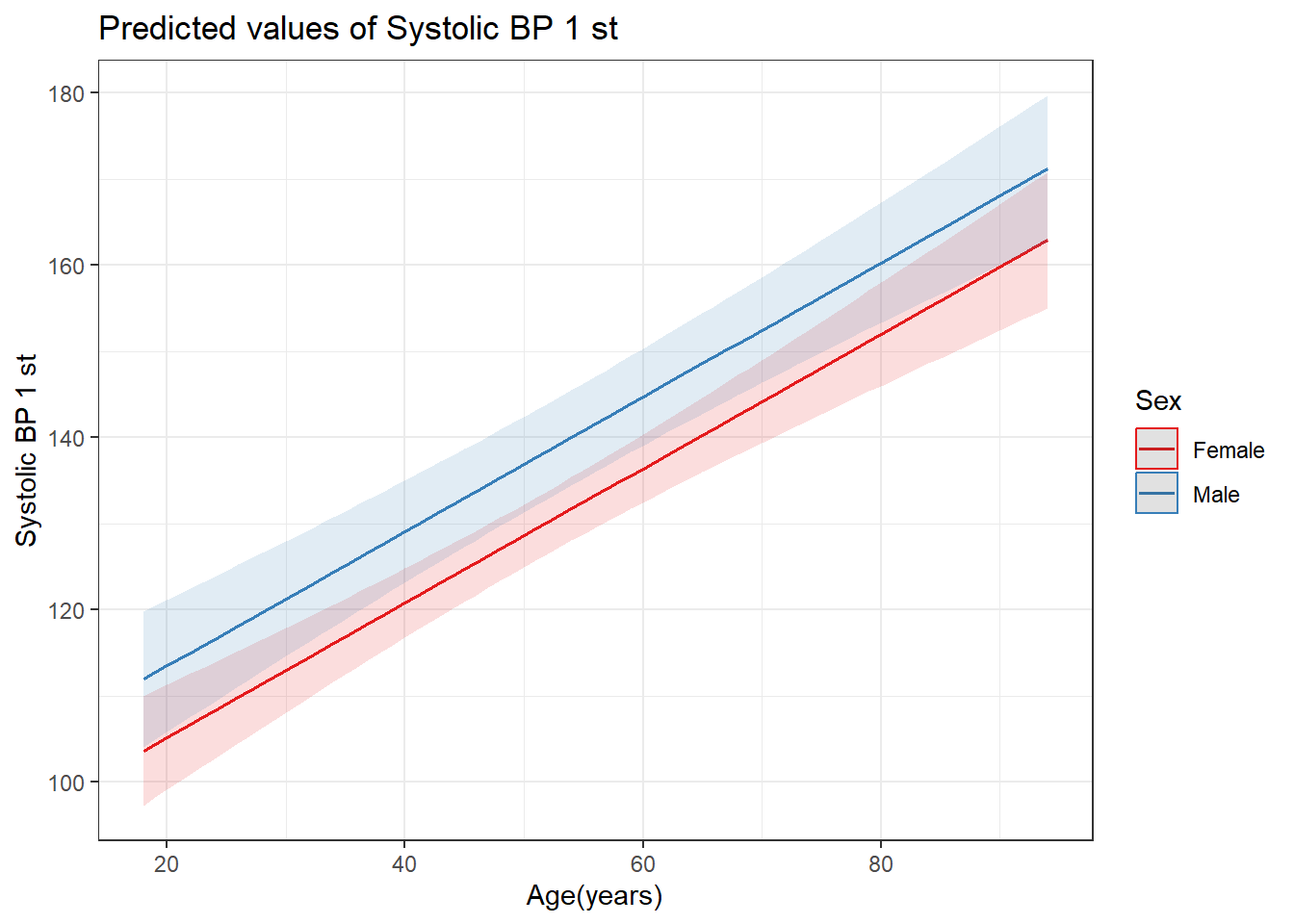
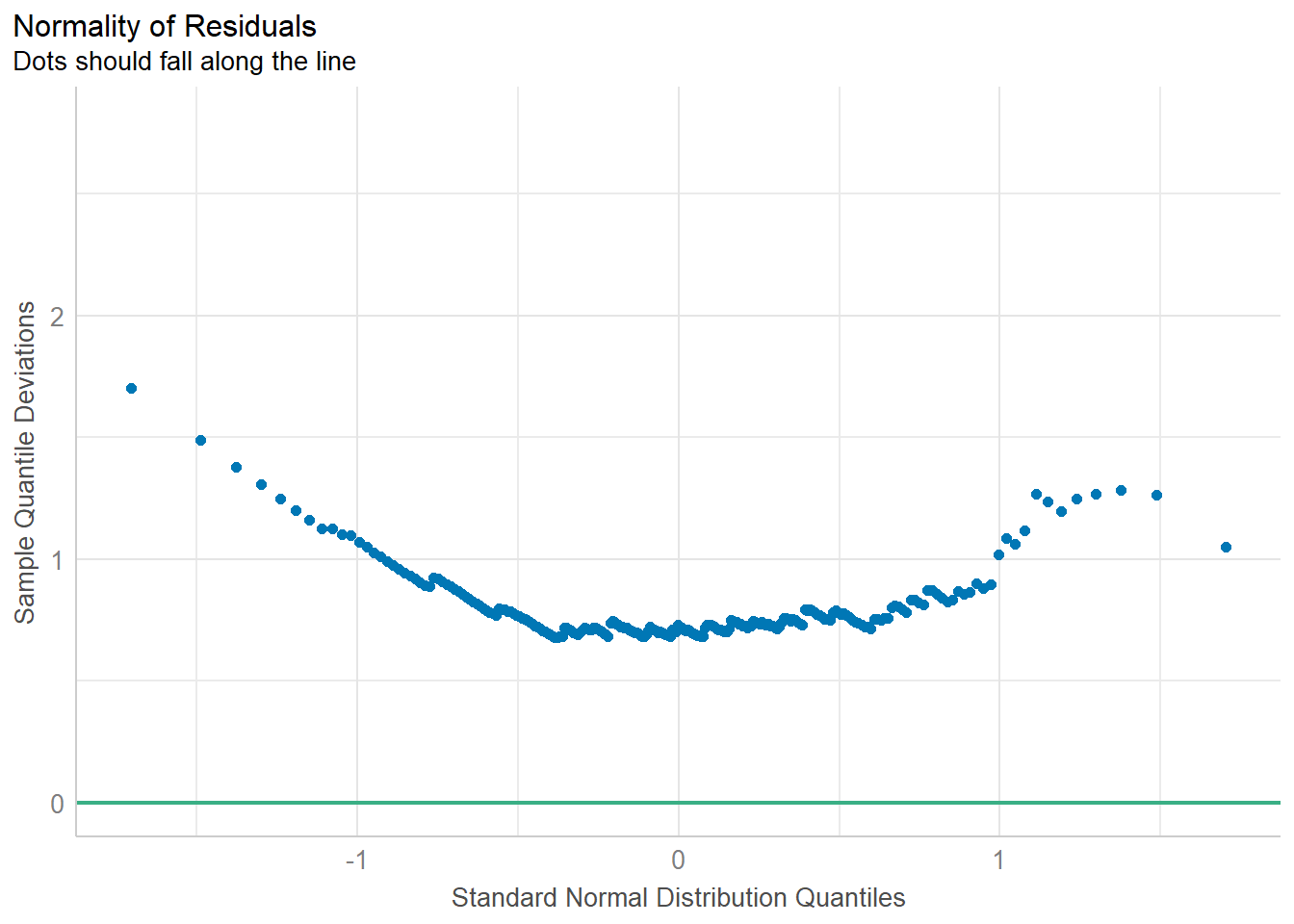
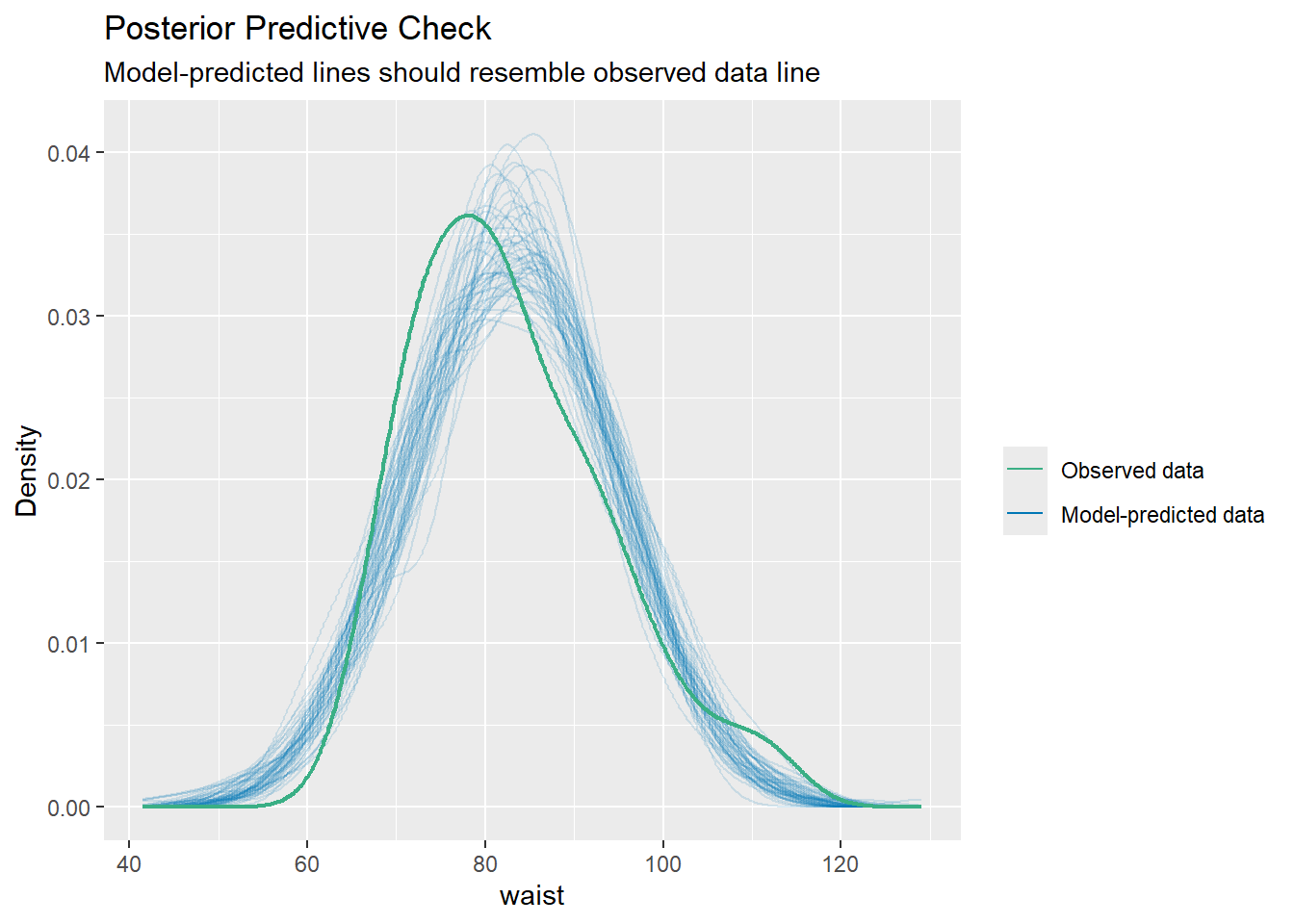
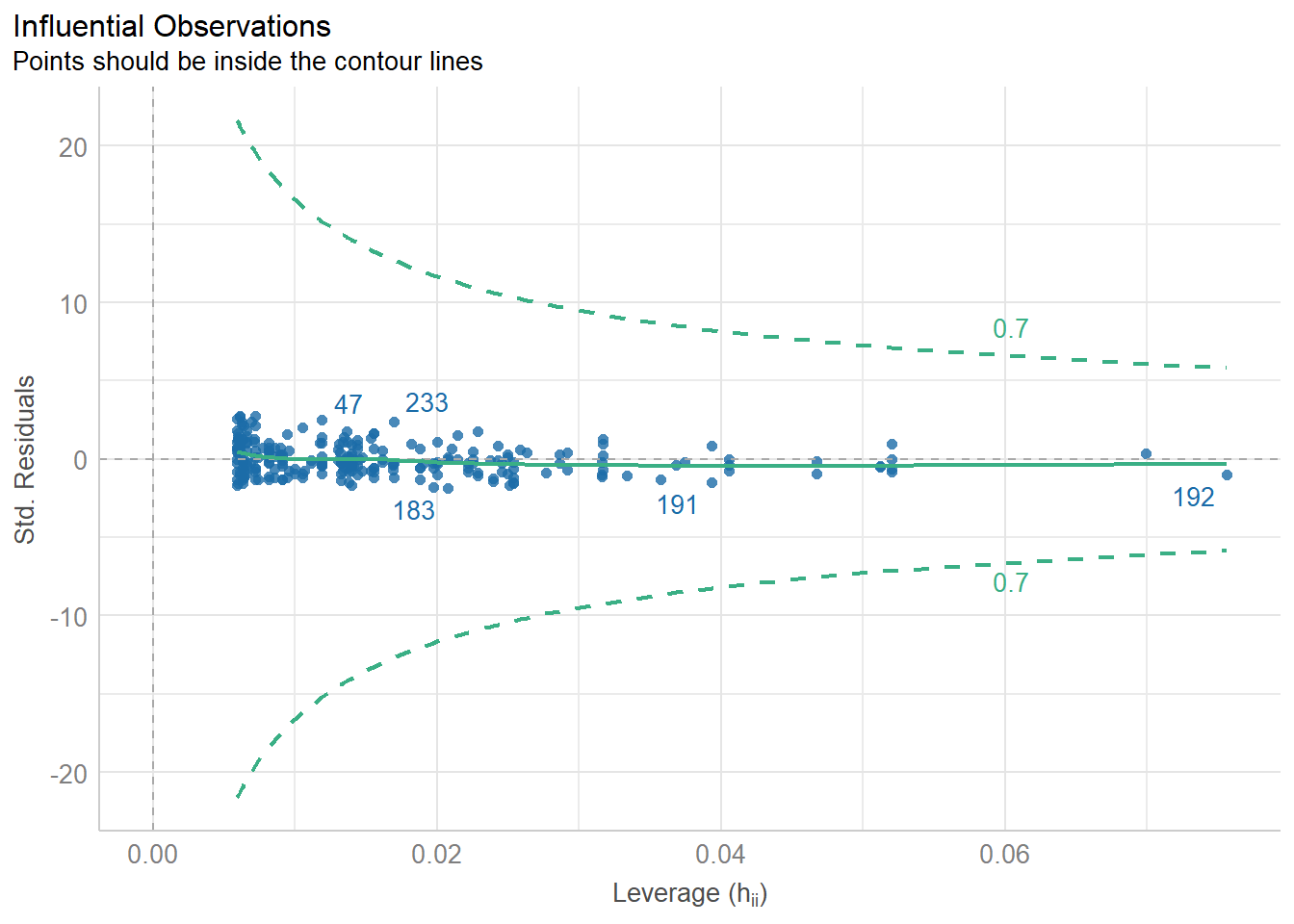
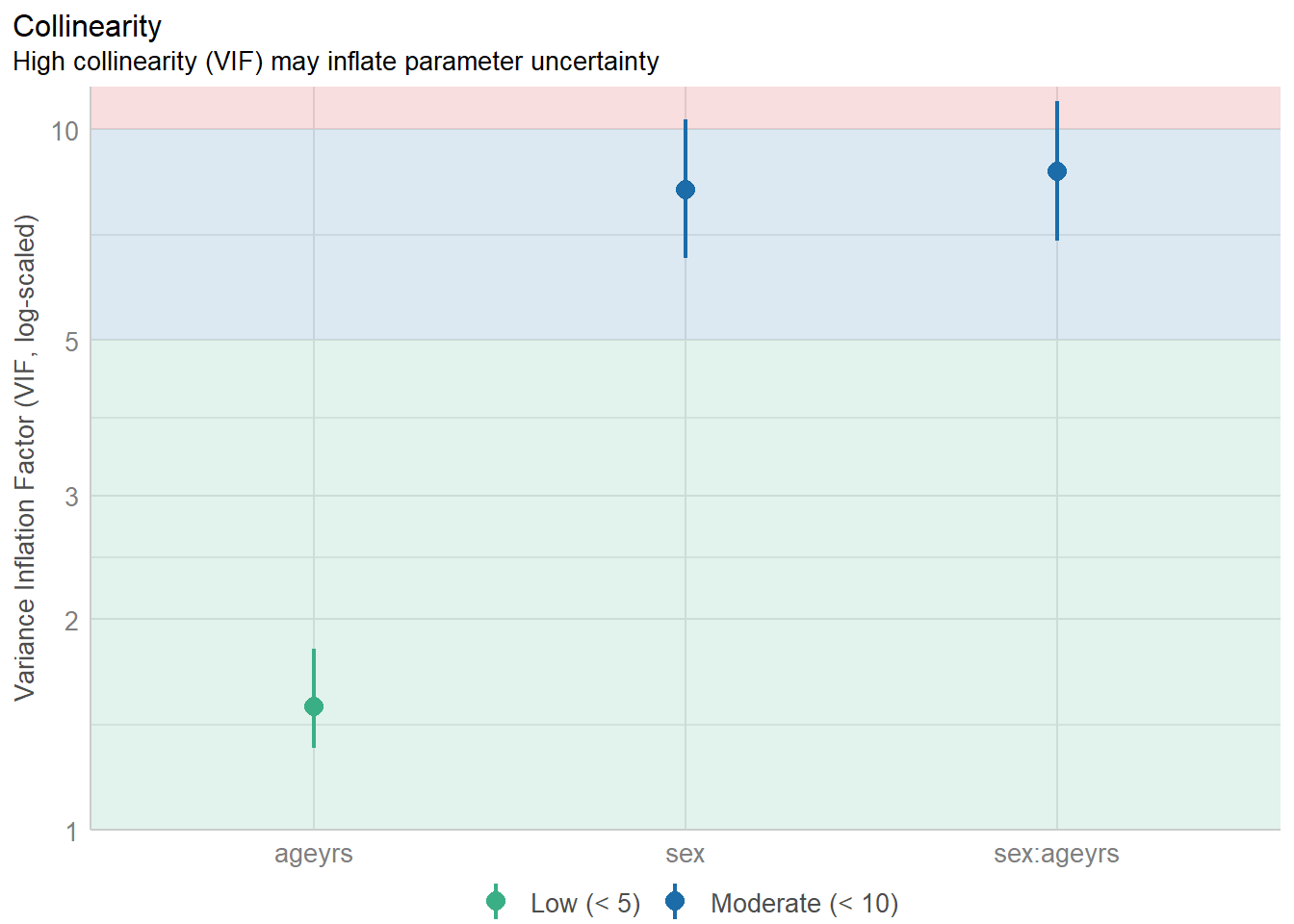
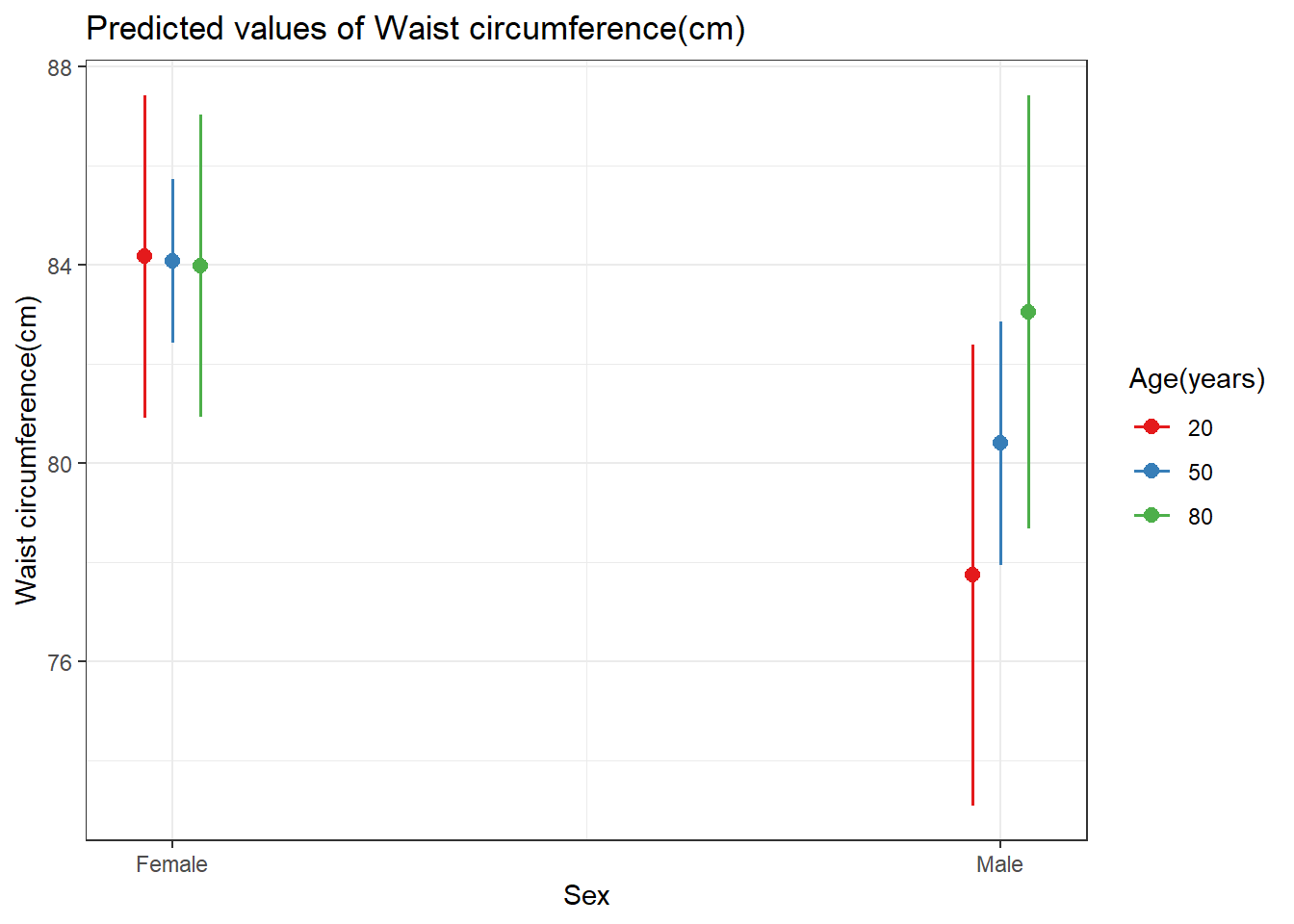
Importing data
Code
library (tidyverse)<- :: read_xlsx (path = "C:/Dataset/hptdata.xlsx" ) %>% :: clean_names () %>% mutate (sex = factor (sex),educ = factor (levels = c ("None" , "Primary" , "JHS/Form 4" , "SHS/Secondary" , "Tertiary" )hpt = case_when (>= 140 | syst2 >= 140 | diast1 >= 90 | diast2 >= 90 ) ~ "Yes" ,< 140 & syst2 < 140 & diast1 < 90 & diast2 < 90 ) ~ "No" %>% factor ()
Labelling data
Code
<- %>% :: set_variable_labels (sid = "Study ID" ,ageyrs = "Age (years)" ,sex = "Sex" ,educ = "Educational level" ,wgt = "Body Weight" ,waist = "Waist circumference (cm)" ,hgt = "Height (cm)" ,syst1 = "Systolic BP 1st" ,diast1 = "Diastolic BP 1st" ,syst2 = "Systolic BP 2nd" ,diast2 = "Diastolic BP 2nd" ,recdate = "Record date" ,hpt = "Hypertension"
Summarizing data
Code
%>% :: dfSummary (graph.col = F, labels.col = F)
Data Frame Summary
df_hpt
Dimensions: 250 x 13
Duplicates: 0
-----------------------------------------------------------------------------------------------
No Variable Stats / Values Freqs (% of Valid) Valid Missing
---- ------------------- --------------------------- --------------------- ---------- ---------
1 sid 1. H001 1 ( 0.4%) 250 0
[character] 2. H002 1 ( 0.4%) (100.0%) (0.0%)
3. H003 1 ( 0.4%)
4. H004 1 ( 0.4%)
5. H005 1 ( 0.4%)
6. H006 1 ( 0.4%)
7. H007 1 ( 0.4%)
8. H008 1 ( 0.4%)
9. H009 1 ( 0.4%)
10. H010 1 ( 0.4%)
[ 240 others ] 240 (96.0%)
2 ageyrs Mean (sd) : 51.4 (18.7) 61 distinct values 250 0
[numeric] min < med < max: (100.0%) (0.0%)
18 < 51 < 94
IQR (CV) : 28 (0.4)
3 sex 1. Female 170 (68.8%) 247 3
[factor] 2. Male 77 (31.2%) (98.8%) (1.2%)
4 educ 1. None 112 (45.0%) 249 1
[factor] 2. Primary 34 (13.7%) (99.6%) (0.4%)
3. JHS/Form 4 84 (33.7%)
4. SHS/Secondary 15 ( 6.0%)
5. Tertiary 4 ( 1.6%)
5 wgt Mean (sd) : 59.8 (12.8) 182 distinct values 246 4
[numeric] min < med < max: (98.4%) (1.6%)
35.5 < 57.6 < 105.2
IQR (CV) : 17.3 (0.2)
6 waist Mean (sd) : 83 (11) 49 distinct values 249 1
[numeric] min < med < max: (99.6%) (0.4%)
64 < 81 < 114
IQR (CV) : 15 (0.1)
7 hgt Mean (sd) : 160.7 (12) 47 distinct values 248 2
[numeric] min < med < max: (99.2%) (0.8%)
67 < 161 < 184
IQR (CV) : 10.5 (0.1)
8 syst1 Mean (sd) : 132.3 (27.8) 98 distinct values 250 0
[numeric] min < med < max: (100.0%) (0.0%)
74 < 128 < 225
IQR (CV) : 35.8 (0.2)
9 diast1 Mean (sd) : 78.2 (16.4) 64 distinct values 247 3
[numeric] min < med < max: (98.8%) (1.2%)
49 < 76 < 160
IQR (CV) : 20 (0.2)
10 syst2 Mean (sd) : 131.6 (27.2) 94 distinct values 250 0
[numeric] min < med < max: (100.0%) (0.0%)
84 < 127 < 232
IQR (CV) : 36 (0.2)
11 diast2 Mean (sd) : 77.9 (15.8) 63 distinct values 247 3
[numeric] min < med < max: (98.8%) (1.2%)
50 < 76 < 150
IQR (CV) : 18.5 (0.2)
12 recdate min : 2012-02-12 250 distinct values 250 0
[POSIXct, POSIXt] med : 2012-06-15 12:00:00 (100.0%) (0.0%)
max : 2012-10-18
range : 8m 6d
13 hpt 1. No 146 (58.6%) 249 1
[factor] 2. Yes 103 (41.4%) (99.6%) (0.4%)
-----------------------------------------------------------------------------------------------
Viewing missing data pattern
Code
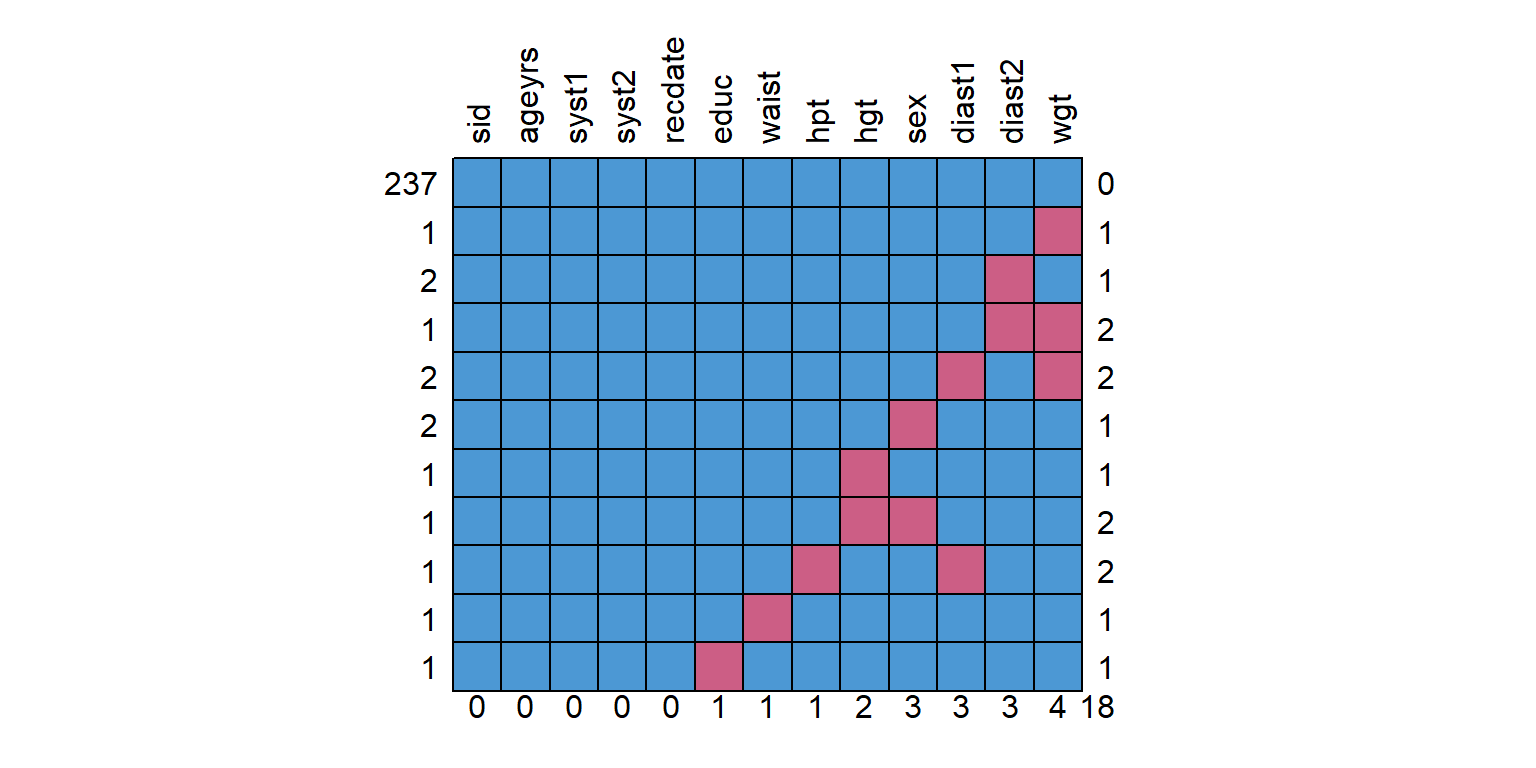
%>% mice:: md.pattern (rotate.names = T)
sid ageyrs syst1 syst2 recdate educ waist hpt hgt sex diast1 diast2 wgt
237 1 1 1 1 1 1 1 1 1 1 1 1 1 0
1 1 1 1 1 1 1 1 1 1 1 1 1 0 1
2 1 1 1 1 1 1 1 1 1 1 1 0 1 1
1 1 1 1 1 1 1 1 1 1 1 1 0 0 2
2 1 1 1 1 1 1 1 1 1 1 0 1 0 2
2 1 1 1 1 1 1 1 1 1 0 1 1 1 1
1 1 1 1 1 1 1 1 1 0 1 1 1 1 1
1 1 1 1 1 1 1 1 1 0 0 1 1 1 2
1 1 1 1 1 1 1 1 0 1 1 0 1 1 2
1 1 1 1 1 1 1 0 1 1 1 1 1 1 1
1 1 1 1 1 1 0 1 1 1 1 1 1 1 1
0 0 0 0 0 1 1 1 2 3 3 3 4 18
Code
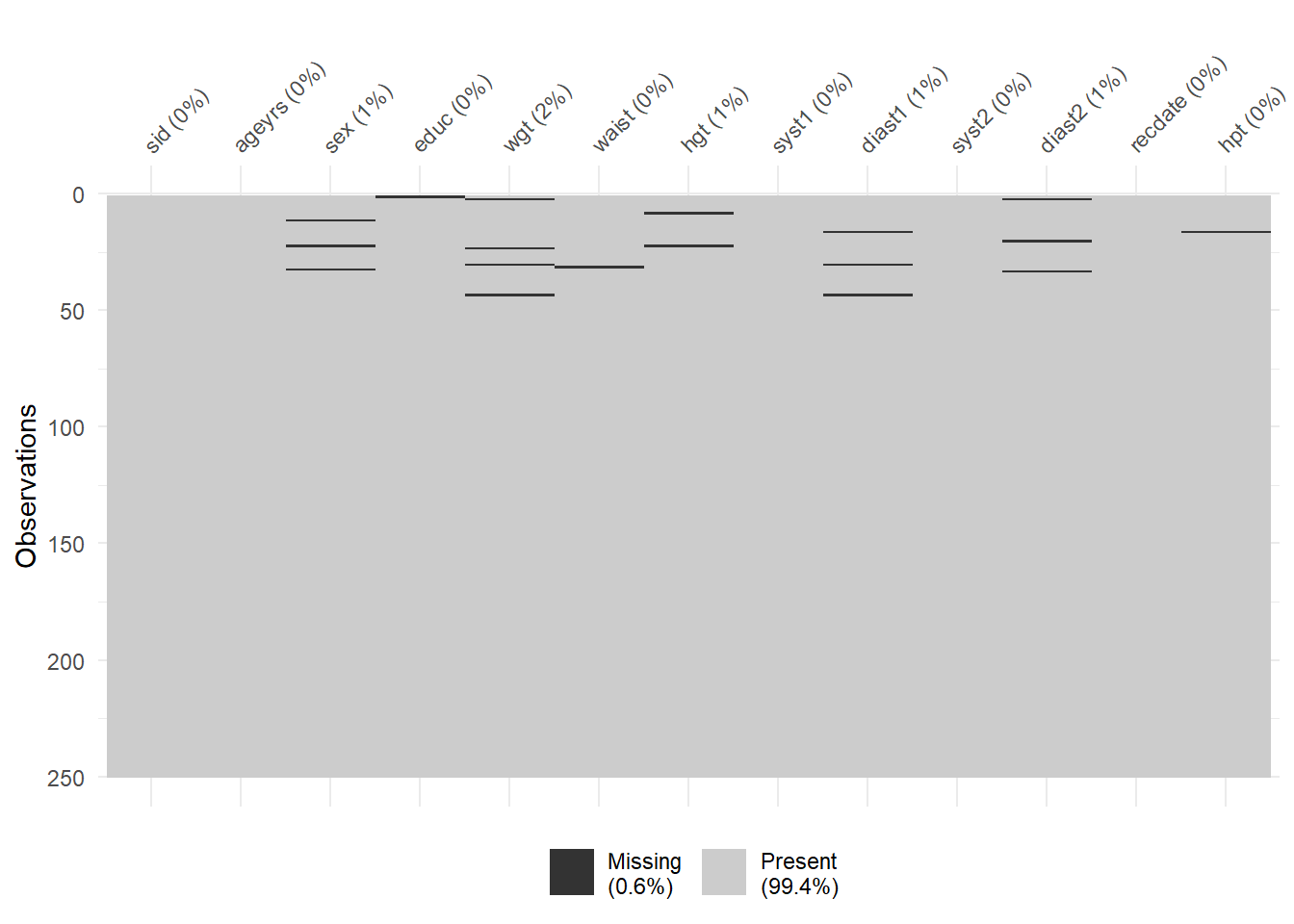
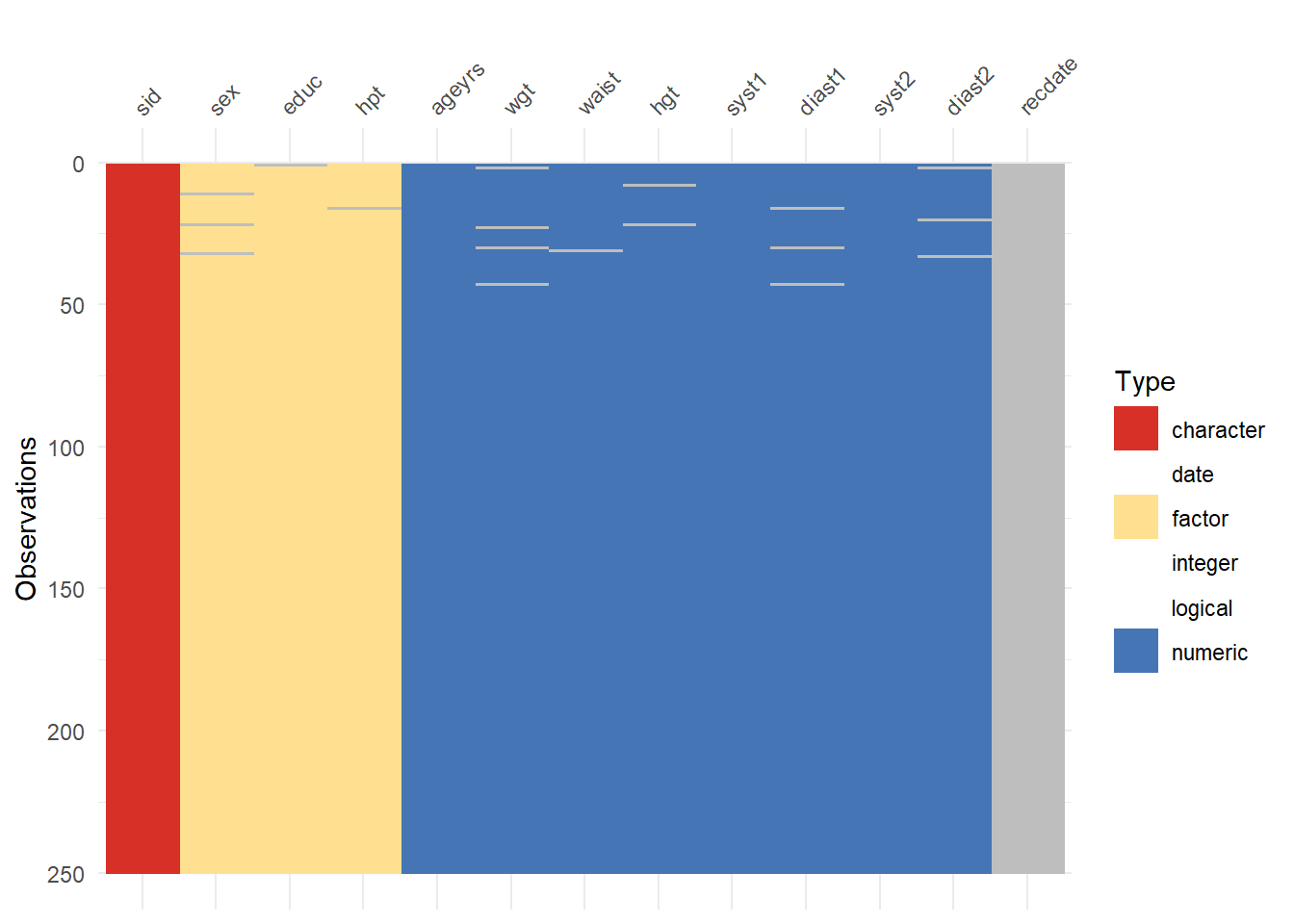
%>% :: vis_miss ()%>% visdat:: vis_dat (palette = "cb_safe" )
Regression with mice imputation
Code
:: theme_gtsummary_compact ()# Input the data <- %>% select (hpt, ageyrs, sex, waist, educ, wgt, hgt) %>% :: mice (maxit = 20 , m = 5 ,printFlag = F)# Visualize the 3rd og the 5 data sets created :: complete (imputed_data, 4 ) %>% head () %>% :: gt () %>% :: opt_stylize (style = 3 , color = "blue" , add_row_striping = TRUE )
Yes
84
Female
83
None
53.7
153
Yes
70
Female
80
None
42.6
149
Yes
55
Female
113
None
97.6
162
No
57
Male
76
JHS/Form 4
59.2
172
No
54
Male
78
JHS/Form 4
58.2
168
No
19
Female
71
SHS/Secondary
54.9
162
Code
# Create univariate table for original data set <- %>% :: tbl_uvregression (include = c (ageyrs, sex, waist, educ, wgt, hgt),y = hpt,method = glm,method.args = family (binomial),exponentiate= TRUE ,pvalue_fun = function (x) gtsummary:: style_pvalue (x, digits = 3 )%>% :: bold_labels () %>% :: bold_p ()
Age (years)
249
1.06
1.04, 1.08
<0.001
Sex
246
Female
—
—
Male
1.09
0.63, 1.87
0.765
Waist circumference (cm)
248
1.02
1.00, 1.04
0.099
Educational level
248
None
—
—
Primary
0.23
0.09, 0.56
0.002
JHS/Form 4
0.43
0.24, 0.77
0.005
SHS/Secondary
0.32
0.08, 0.98
0.060
Tertiary
2.60
0.32, 53.4
0.414
Body Weight
245
0.98
0.96, 1.00
0.120
Height (cm)
247
0.98
0.96, 1.00
0.103
Code
# Build the model <- with (imputed_data, glm (hpt ~ ageyrs + sex + waist + educ + wgt + hgt, family = "binomial" )# present beautiful table with gtsummary <- %>% :: tbl_regression (exponentiate= TRUE ,pvalue_fun = function (x) gtsummary:: style_pvalue (x, digits = 3 )%>% :: bold_labels () %>% :: bold_p ()
Age (years)
1.06
1.04, 1.09
<0.001
Sex
Female
—
—
Male
1.29
0.60, 2.78
0.507
Waist circumference (cm)
1.03
0.98, 1.09
0.242
Educational level
None
—
—
Primary
0.52
0.18, 1.47
0.217
JHS/Form 4
0.86
0.41, 1.79
0.683
SHS/Secondary
0.85
0.21, 3.49
0.827
Tertiary
11.1
0.89, 140
0.062
Body Weight
1.00
0.95, 1.05
0.861
Height (cm)
0.99
0.96, 1.02
0.438
Code
# Combine tables :: tbl_merge (tbls = list (tbl1, tbl2), tab_spanner = c ("**Univariate**" , "**Multivariate**" )
N OR 95% CI p-value OR 95% CI p-value
Age (years)
249
1.06
1.04, 1.08
<0.001
1.06
1.04, 1.09
<0.001
Sex
246
Female
—
—
—
—
Male
1.09
0.63, 1.87
0.765
1.29
0.60, 2.78
0.507
Waist circumference (cm)
248
1.02
1.00, 1.04
0.099
1.03
0.98, 1.09
0.242
Educational level
248
None
—
—
—
—
Primary
0.23
0.09, 0.56
0.002
0.52
0.18, 1.47
0.217
JHS/Form 4
0.43
0.24, 0.77
0.005
0.86
0.41, 1.79
0.683
SHS/Secondary
0.32
0.08, 0.98
0.060
0.85
0.21, 3.49
0.827
Tertiary
2.60
0.32, 53.4
0.414
11.1
0.89, 140
0.062
Body Weight
245
0.98
0.96, 1.00
0.120
1.00
0.95, 1.05
0.861
Height (cm)
247
0.98
0.96, 1.00
0.103
0.99
0.96, 1.02
0.438